Arabidopsis kinesin KP1 specifically interacts with VDAC3, a mitochondrial protein, and regulates respiration during seed germination at low temperature
- PMID: 21406623
- PMCID: PMC3082256
- DOI: 10.1105/tpc.110.082420
Arabidopsis kinesin KP1 specifically interacts with VDAC3, a mitochondrial protein, and regulates respiration during seed germination at low temperature
Abstract
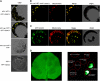
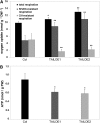
The involvement of cytoskeleton-related proteins in regulating mitochondrial respiration has been revealed in mammalian cells. However, it is unclear if there is a relationship between the microtubule-based motor protein kinesin and mitochondrial respiration. In this research, we demonstrate that a plant-specific kinesin, Kinesin-like protein 1 (KP1; At KIN14 h), is involved in respiratory regulation during seed germination at a low temperature. Using in vitro biochemical methods and in vivo transgenic cell observations, we demonstrate that KP1 is able to localize to mitochondria via its tail domain (C terminus) and specifically interacts with a mitochondrial outer membrane protein, voltage-dependent anion channel 3 (VDAC3). Targeting of the KP1-tail to mitochondria is dependent on the presence of VDAC3. When grown at 4° C, KP1 dominant-negative mutants (TAILOEs) and vdac3 mutants exhibited a higher seed germination frequency. All germinating seeds of the kp1 and vdac3 mutants had increased oxygen consumption; the respiration balance between the cytochrome pathway and the alternative oxidase pathway was disrupted, and the ATP level was reduced. We conclude that the plant-specific kinesin, KP1, specifically interacts with VDAC3 on the mitochondrial outer membrane and that both KP1 and VDAC3 regulate aerobic respiration during seed germination at low temperature.
Figures









References
-
- Benz R. (1994). Permeation of hydrophilic solutes through mitochondrial outer membranes: Review on mitochondrial porins. Biochim. Biophys. Acta 1197: 167–196 - PubMed
-
- Bond D.R., Mester T., Nesbø C.L., Izquierdo-Lopez A.V., Collart F.L., Lovley D.R. (2005). Characterization of citrate synthase from Geobacter sulfurreducens and evidence for a family of citrate synthases similar to those of eukaryotes throughout the Geobacteraceae. Appl. Environ. Microbiol. 71: 3858–3865 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

