High-resolution identity by descent mapping uncovers the genetic basis for blood pressure differences between spontaneously hypertensive rat lines
- PMID: 21406686
- PMCID: PMC3116070
- DOI: 10.1161/CIRCGENETICS.110.958934
High-resolution identity by descent mapping uncovers the genetic basis for blood pressure differences between spontaneously hypertensive rat lines
Abstract
Background: The recent development of a large panel of genome-wide single nucleotide polymorphisms (SNPs) provides the opportunity to examine genetic relationships between distinct SHR lines that share hypertension but differ in their susceptibility to hypertensive end-organ disease.
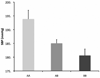
Methods and results: We compared genotypes at nearly 10,000 SNPs obtained for the hypertension end-organ injury-susceptible spontaneously hypertensive rat (SHR)-A3 (SHRSP, SHR-stroke prone) line and the injury-resistant SHR-B2 line. This revealed that that the 2 lines were genetically identical by descent (IBD) across 86.6% of the genome. Areas of the genome that were not IBD were distributed across 19 of the 20 autosomes and the X chromosome. A block structure of non-IBD comprising a total of 121 haplotype blocks was formed by clustering of SNPs inherited from different ancestors. To test the null hypothesis that distinct SHR lines share a common set of hypertension susceptibility alleles, we compared blood pressure in adult SHR animals from both lines and their F1 and F2 progeny using telemetry. In 16- to 18-week-old animals fed a normal diet, systolic blood pressure (SBP, mm Hg) in SHR-A3 was 205.7 ± 3.86 (mean ± SEM, n = 26), whereas in similar SHR-B2 animals, SBP was 186.7 ± 2.53 (n = 20). In F1 and F2 animals, SBP was 188.2 ± 4.23 (n = 19) and 185.6 ± 1.1 (n = 211), respectively (P<10(-6), ANOVA). To identify non-IBD haplotype blocks contributing to blood pressure differences between these SHR lines, we developed a high-throughput SNP genotyping system to genotype SNPs marking non-IBD blocks. We mapped a single non-IBD block on chromosome 17 extending over <10 Mb, at which SHR-A3 alleles significantly elevate blood pressure compared with SHR-B2.
Conclusions: Thus hypertension in SHR-A3 and -B2 appears to arise from an overlapping set of susceptibility alleles, with SHR-A3 possessing an additional hypertension locus that contributes to further increase blood pressure.
Conflict of interest statement
Figures




References
-
- Okamoto K, Yamori Y, Nosaka S, Ooshima A, Hazama F. Studies on hypertension in spontaneously hypertensive rats. Clin Sci Mol Med Suppl. 1973;(45 Suppl 1) 11s-14. - PubMed
-
- Okamoto K, Yamori Y, Nagaoka A. Establishment of the stroke-prone spontaneously hypertensive rat. Circ. Res. 1974;34/35:I-143–I-153.
-
- Nagaoka A, Iwatsuka H, Suzuoki Z, Okamoto K. Genetic predisposition to stroke in spontaneously hypertensive rats. Am J Physiol. 1976;230:1354–1359. - PubMed
-
- Gigante B, Rubattu S, Stanzione R, Lombardi A, Baldi A, Baldi F, Volpe M. Contribution of genetic factors to renal lesions in the stroke-prone spontaneously hypertensive rat. Hypertension. 2003;42:702–706. - PubMed
-
- Hilbert P, Lindpaintner K, Beckmann JS, Serikawa T, Soubrier F, Dubay C, Cartwright P, De Gouyon B, Julier C, Takahasi S, Vincent M, Ganten D, Georges M, Lathrop GM. Chromosomal mapping of two genetic loci associated with blood-pressure regulation in hereditary hypertensive rats. Nature. 1991;353:521–529. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

