Damage-induced localized hypermutability
- PMID: 21406975
- PMCID: PMC3100884
- DOI: 10.4161/cc.10.7.15319
Damage-induced localized hypermutability
Abstract
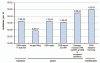
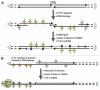
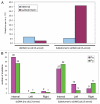
Genome instability continuously presents perils of cancer, genetic disease and death of a cell or an organism. At the same time, it provides for genome plasticity that is essential for development and evolution. We address here the genome instability confined to a small fraction of DNA adjacent to free DNA ends at uncapped telomeres and double-strand breaks. We found that budding yeast cells can tolerate nearly 20 kilobase regions of subtelomeric single-strand DNA that contain multiple UV-damaged nucleotides. During restoration to the double-strand state, multiple mutations are generated by error-prone translesion synthesis. Genome-wide sequencing demonstrated that multiple regions of damage-induced localized hypermutability can be tolerated, which leads to the simultaneous appearance of multiple mutation clusters in the genomes of UV- irradiated cells. High multiplicity and density of mutations suggest that this novel form of genome instability may play significant roles in generating new alleles for evolutionary selection as well as in the incidence of cancer and genetic disease.
Figures


References
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA repair and mutagenesis. Washington: ASM Press; 2006.
-
- Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
