Miniproteins as phage display-scaffolds for clinical applications
- PMID: 21407148
- PMCID: PMC6259850
- DOI: 10.3390/molecules16032467
Miniproteins as phage display-scaffolds for clinical applications
Abstract
Miniproteins are currently developed as alternative, non-immunoglobin proteins for the generation of novel binding motifs. Miniproteins are rigid scaffolds that are stabilised by alpha-helices, beta-sheets and disulfide-constrained secondary structural elements. They are tolerant to multiple amino acid substitutions, which allow for the integration of a randomised affinity function into the stably folded framework. These properties classify miniprotein scaffolds as promising tools for lead structure generation using phage display technologies. Owing to their high enzymatic resistance and structural stability, miniproteins are ideal templates to display binding epitopes for medical applications in vivo. This review summarises the characteristics and the engineering of miniproteins as a novel class of scaffolds to generate of alternative binding agents using phage display screening. Moreover, recent developments for therapeutic and especially diagnostic applications of miniproteins are reviewed.
Figures
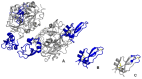
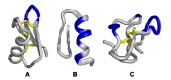


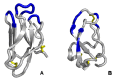
Similar articles
-
Miniprotein Design: Past, Present, and Prospects.Acc Chem Res. 2017 Sep 19;50(9):2085-2092. doi: 10.1021/acs.accounts.7b00186. Epub 2017 Aug 23. Acc Chem Res. 2017. PMID: 28832117
-
Engineered cystine-knot miniproteins for diagnostic applications.Expert Rev Mol Diagn. 2010 Apr;10(3):361-8. doi: 10.1586/erm.10.15. Expert Rev Mol Diagn. 2010. PMID: 20370592 Review.
-
Alternative binding proteins: biological activity and therapeutic potential of cystine-knot miniproteins.FEBS J. 2008 Jun;275(11):2684-90. doi: 10.1111/j.1742-4658.2008.06440.x. Epub 2008 Apr 24. FEBS J. 2008. PMID: 18435757 Review.
-
Novel miniproteins engineered by the transfer of active sites to small natural scaffolds.Biopolymers. 1998;47(1):93-100. doi: 10.1002/(SICI)1097-0282(1998)47:1<93::AID-BIP10>3.0.CO;2-H. Biopolymers. 1998. PMID: 9692330 Review.
-
Determinants of Developability and Evolvability of Synthetic Miniproteins as Ligand Scaffolds.J Mol Biol. 2023 Dec 15;435(24):168339. doi: 10.1016/j.jmb.2023.168339. Epub 2023 Nov 3. J Mol Biol. 2023. PMID: 37923119 Free PMC article.
Cited by
-
Computational Structural and Functional Analyses of ORF10 in Novel Coronavirus SARS-CoV-2 Variants to Understand Evolutionary Dynamics.Evol Bioinform Online. 2022 Jul 7;18:11769343221108218. doi: 10.1177/11769343221108218. eCollection 2022. Evol Bioinform Online. 2022. PMID: 35909986 Free PMC article.
-
Noncompetitive Allosteric Antagonism of Death Receptor 5 by a Synthetic Affibody Ligand.Biochemistry. 2020 Oct 13;59(40):3856-3868. doi: 10.1021/acs.biochem.0c00529. Epub 2020 Sep 30. Biochemistry. 2020. PMID: 32941010 Free PMC article.
-
Beyond Antibodies as Binding Partners: The Role of Antibody Mimetics in Bioanalysis.Annu Rev Anal Chem (Palo Alto Calif). 2017 Jun 12;10(1):293-320. doi: 10.1146/annurev-anchem-061516-045205. Epub 2017 Mar 24. Annu Rev Anal Chem (Palo Alto Calif). 2017. PMID: 28375702 Free PMC article. Review.
-
Discovery and Characterization of a Potent Interleukin-6 Binding Peptide with Neutralizing Activity In Vivo.PLoS One. 2015 Nov 10;10(11):e0141330. doi: 10.1371/journal.pone.0141330. eCollection 2015. PLoS One. 2015. PMID: 26555695 Free PMC article.
-
Beyond antibody engineering: directed evolution of alternative binding scaffolds and enzymes using yeast surface display.Microb Cell Fact. 2018 Feb 26;17(1):32. doi: 10.1186/s12934-018-0881-3. Microb Cell Fact. 2018. PMID: 29482656 Free PMC article. Review.
References
-
- Holliger P., Hudson P.J. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 2005;23:1126–1136. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

