Contribution of CgPDR1-regulated genes in enhanced virulence of azole-resistant Candida glabrata
- PMID: 21408004
- PMCID: PMC3052359
- DOI: 10.1371/journal.pone.0017589
Contribution of CgPDR1-regulated genes in enhanced virulence of azole-resistant Candida glabrata
Abstract
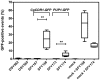
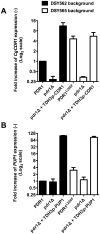
In Candida glabrata, the transcription factor CgPdr1 is involved in resistance to azole antifungals via upregulation of ATP binding cassette (ABC)-transporter genes including at least CgCDR1, CgCDR2 and CgSNQ2. A high diversity of GOF (gain-of-function) mutations in CgPDR1 exists for the upregulation of ABC-transporters. These mutations enhance C. glabrata virulence in animal models, thus indicating that CgPDR1 might regulate the expression of yet unidentified virulence factors. We hypothesized that CgPdr1-dependent virulence factor(s) should be commonly regulated by all GOF mutations in CgPDR1. As deduced from transcript profiling with microarrays, a high number of genes (up to 385) were differentially regulated by a selected number (7) of GOF mutations expressed in the same genetic background. Surprisingly, the transcriptional profiles resulting from expression of GOF mutations showed minimal overlap in co-regulated genes. Only two genes, CgCDR1 and PUP1 (for PDR1 upregulated and encoding a mitochondrial protein), were commonly upregulated by all tested GOFs. While both genes mediated azole resistance, although to different extents, their deletions in an azole-resistant isolate led to a reduction of virulence and decreased tissue burden as compared to clinical parents. As expected from their role in C. glabrata virulence, the two genes were expressed as well in vitro and in vivo. The individual overexpression of these two genes in a CgPDR1-independent manner could partially restore phenotypes obtained in clinical isolates. These data therefore demonstrate that at least these two CgPDR1-dependent and -upregulated genes contribute to the enhanced virulence of C. glabrata that acquired azole resistance.
Conflict of interest statement
Figures







References
-
- Kaur R, Domergue R, Zupancic ML, Cormack BP. A yeast by any other name: Candida glabrata and its interaction with the host. Curr Opin Microbiol. 2005;8:378–384. - PubMed
-
- Ruhnke M. Epidemiology of Candida albicans infections and role of non-Candida albicans yeasts. Curr Drug Targets. 2006;7:495–504. - PubMed
-
- Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Barton R, et al. Geographic variation in the frequency of isolation and fluconazole and voriconazole susceptibilities of Candida glabrata: an assessment from the ARTEMIS DISK Global Antifungal Surveillance Program. Diagn Microbiol Infect Dis. 2010;67:162–171. - PubMed
-
- Sanglard D, Odds FC. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect Dis. 2002;2:73–85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

