A bow-tie genetic architecture for morphogenesis suggested by a genome-wide RNAi screen in Caenorhabditis elegans
- PMID: 21408209
- PMCID: PMC3048373
- DOI: 10.1371/journal.pgen.1002010
A bow-tie genetic architecture for morphogenesis suggested by a genome-wide RNAi screen in Caenorhabditis elegans
Abstract
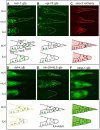
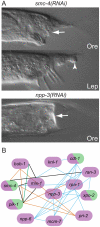
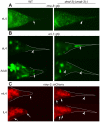
During animal development, cellular morphogenesis plays a fundamental role in determining the shape and function of tissues and organs. Identifying the components that regulate and drive morphogenesis is thus a major goal of developmental biology. The four-celled tip of the Caenorhabditis elegans male tail is a simple but powerful model for studying the mechanism of morphogenesis and its spatiotemporal regulation. Here, through a genome-wide post-embryonic RNAi-feeding screen, we identified 212 components that regulate or participate in male tail tip morphogenesis. We constructed a working hypothesis for a gene regulatory network of tail tip morphogenesis. We found regulatory roles for the posterior Hox genes nob-1 and php-3, the TGF-β pathway, nuclear hormone receptors (e.g. nhr-25), the heterochronic gene blmp-1, and the GATA transcription factors egl-18 and elt-6. The majority of the pathways converge at dmd-3 and mab-3. In addition, nhr-25 and dmd-3/mab-3 regulate each others' expression, thus placing these three genes at the center of a complex regulatory network. We also show that dmd-3 and mab-3 negatively regulate other signaling pathways and affect downstream cellular processes such as vesicular trafficking (e.g. arl-1, rme-8) and rearrangement of the cytoskeleton (e.g. cdc-42, nmy-1, and nmy-2). Based on these data, we suggest that male tail tip morphogenesis is governed by a gene regulatory network with a bow-tie architecture.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Tissue-specific RNA-seq defines genes governing male tail tip morphogenesis in C. elegans.Development. 2024 Sep 15;151(18):dev202787. doi: 10.1242/dev.202787. Epub 2024 Sep 24. Development. 2024. PMID: 39253748
-
Novel gain-of-function alleles demonstrate a role for the heterochronic gene lin-41 in C. elegans male tail tip morphogenesis.Dev Biol. 2006 Sep 1;297(1):74-86. doi: 10.1016/j.ydbio.2006.04.472. Epub 2006 May 6. Dev Biol. 2006. PMID: 16806150
-
dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans.Development. 2008 Aug;135(14):2373-82. doi: 10.1242/dev.017046. Epub 2008 Jun 11. Development. 2008. PMID: 18550714 Free PMC article.
-
Genes that shape the course of ageing.Trends Endocrinol Metab. 2003 Oct;14(8):345-7. doi: 10.1016/j.tem.2003.08.003. Trends Endocrinol Metab. 2003. PMID: 14516929 Review.
-
Networks in Caenorhabditis elegans.Curr Opin Genet Dev. 2011 Dec;21(6):787-98. doi: 10.1016/j.gde.2011.10.003. Epub 2011 Nov 4. Curr Opin Genet Dev. 2011. PMID: 22054717 Review.
Cited by
-
Collaborative regulation of development but independent control of metabolism by two epidermis-specific transcription factors in Caenorhabditis elegans.J Biol Chem. 2013 Nov 15;288(46):33411-26. doi: 10.1074/jbc.M113.487975. Epub 2013 Oct 6. J Biol Chem. 2013. PMID: 24097988 Free PMC article.
-
BLMP-1/Blimp-1 regulates the spatiotemporal cell migration pattern in C. elegans.PLoS Genet. 2014 Jun 26;10(6):e1004428. doi: 10.1371/journal.pgen.1004428. eCollection 2014 Jun. PLoS Genet. 2014. PMID: 24968003 Free PMC article.
-
Sexually Dimorphic Differentiation of a C. elegans Hub Neuron Is Cell Autonomously Controlled by a Conserved Transcription Factor.Curr Biol. 2017 Jan 23;27(2):199-209. doi: 10.1016/j.cub.2016.11.045. Epub 2017 Jan 5. Curr Biol. 2017. PMID: 28065609 Free PMC article.
-
The genetic causes of convergent evolution.Nat Rev Genet. 2013 Nov;14(11):751-64. doi: 10.1038/nrg3483. Epub 2013 Oct 9. Nat Rev Genet. 2013. PMID: 24105273 Review.
-
Genome-wide tissue-specific gene expression, co-expression and regulation of co-expressed genes in adult nematode Ascaris suum.PLoS Negl Trop Dis. 2014 Feb 6;8(2):e2678. doi: 10.1371/journal.pntd.0002678. eCollection 2014 Feb. PLoS Negl Trop Dis. 2014. PMID: 24516681 Free PMC article.
References
-
- Sulston JE, Albertson DG, Thomson JN. The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev Biol. 1980;78:542–576. - PubMed
-
- Nguyen CQ, Hall DH, Yang Y, Fitch DH. Morphogenesis of the Caenorhabditis elegans male tail tip. Dev Biol. 1999;207:86–106. - PubMed
-
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. - PubMed
-
- Zhao X, Yang Y, Fitch DH, Herman MA. TLP-1 is an asymmetric cell fate determinant that responds to Wnt signals and controls male tail tip morphogenesis in C. elegans. Development. 2002;129:1497–1508. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

