Azacytidine and decitabine induce gene-specific and non-random DNA demethylation in human cancer cell lines
- PMID: 21408221
- PMCID: PMC3049766
- DOI: 10.1371/journal.pone.0017388
Azacytidine and decitabine induce gene-specific and non-random DNA demethylation in human cancer cell lines
Abstract
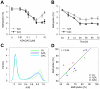
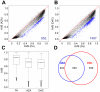
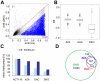
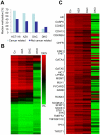
The DNA methyltransferase inhibitors azacytidine and decitabine represent archetypal drugs for epigenetic cancer therapy. To characterize the demethylating activity of azacytidine and decitabine we treated colon cancer and leukemic cells with both drugs and used array-based DNA methylation analysis of more than 14,000 gene promoters. Additionally, drug-induced demethylation was compared to methylation patterns of isogenic colon cancer cells lacking both DNA methyltransferase 1 (DNMT1) and DNMT3B. We show that drug-induced demethylation patterns are highly specific, non-random and reproducible, indicating targeted remethylation of specific loci after replication. Correspondingly, we found that CG dinucleotides within CG islands became preferentially remethylated, indicating a role for DNA sequence context. We also identified a subset of genes that were never demethylated by drug treatment, either in colon cancer or in leukemic cell lines. These demethylation-resistant genes were enriched for Polycomb Repressive Complex 2 components in embryonic stem cells and for transcription factor binding motifs not present in demethylated genes. Our results provide detailed insights into the DNA methylation patterns induced by azacytidine and decitabine and suggest the involvement of complex regulatory mechanisms in drug-induced DNA demethylation.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

