Glycomics-based analysis of chicken red blood cells provides insight into the selectivity of the viral agglutination assay
- PMID: 21410647
- PMCID: PMC3740515
- DOI: 10.1111/j.1742-4658.2011.08096.x
Glycomics-based analysis of chicken red blood cells provides insight into the selectivity of the viral agglutination assay
Abstract
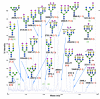
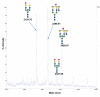
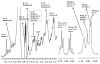
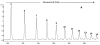
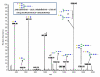
Agglutination of red blood cells (RBCs), including chicken RBCs (cRBCs), has been used extensively to estimate viral titer, to screen glycan-receptor binding preference, and to assess the protective response of vaccines. Although this assay enjoys widespread use, some virus strains do not agglutinate RBCs. To address these underlying issues and to increase the usefulness of cRBCs as tools for studying viruses, such as influenza, we analyzed the cell surface N-glycans of cRBCs. On the basis of the results obtained from complementary analytical strategies, including MS, 1D and 2D-NMR spectroscopy, exoglycosidase digestions, and HPLC profiling, we report the major glycan structures present on cRBCs. By comparing the glycan structures of cBRCs with those of representative human upper respiratory cells, we offer a possible explanation for the fact that certain influenza strains do not agglutinate cRBCs, using specific human-adapted influenza hemagglutinins as examples. Finally, recent understanding of the role of various glycan structures in high affinity binding to influenza hemagglutinins provides context to our findings. These results illustrate that the field of glycomics can provide important information with respect to the experimental systems used to characterize, detect and study viruses.
© 2011 The Authors Journal compilation © 2011 FEBS.
Figures






References
-
- Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, Raman R, Cox NJ, Sasisekharan R, Katz JM, Tumpey TM. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science. 2009;325:484–7. - PMC - PubMed
-
- Boon AC, French AM, Fleming DM, Zambon MC. Detection of influenza a subtypes in community-based surveillance. J Med Virol. 2001;65:163–70. - PubMed
-
- Amano Y, Cheng Q. Detection of influenza virus: traditional approaches and development of biosensors. Anal Bioanal Chem. 2005;381:156–64. - PubMed
-
- Zhang WD, Evans DH. Detection and identification of human influenza viruses by the polymerase chain reaction. J Virol Methods. 1991;33:165–89. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

