Reverse genetic analysis of Ourmiaviruses reveals the nucleolar localization of the coat protein in Nicotiana benthamiana and unusual requirements for virion formation
- PMID: 21411534
- PMCID: PMC3126195
- DOI: 10.1128/JVI.02565-10
Reverse genetic analysis of Ourmiaviruses reveals the nucleolar localization of the coat protein in Nicotiana benthamiana and unusual requirements for virion formation
Abstract
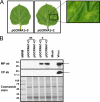
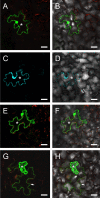
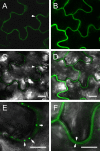
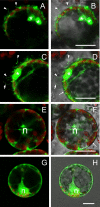
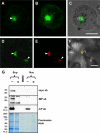
Ourmia melon virus (OuMV) is the type member of the genus Ourmiavirus. These viruses have a trisegmented genome, each part of which encodes a single protein. Ourmiaviruses share a distant similarity with other plant viruses only in their movement proteins (MP), whereas their RNA-dependent RNA polymerase (RdRP) shares features only with fungal viruses of the family Narnaviridae. Thus, ourmiaviruses are in a unique phylogenetic position among existing plant viruses. Here, we developed an agroinoculation system to launch infection in Nicotiana benthamiana plants. Using different combinations of the three segments, we demonstrated that RNA1 is necessary and sufficient for cis-acting replication in the agroinfiltrated area. RNA2 and RNA3, encoding the putative movement protein and the coat protein (CP), respectively, are both necessary for successful systemic infection of N. benthamiana. The CP is dispensable for long-distance transport of the virus through vascular tissues, but its absence prevents efficient systemic infection at the exit sites. Virion formation occurred only when the CP was translated from replication-derived RNA3. Transient expression of a green fluorescent protein-MP (GFP-MP) fusion via agroinfiltration showed that the MP is present in cytoplasmic connections across plant cell walls; in protoplasts the GFP-MP fusion stimulates the formation of tubular protrusions. Expression through agroinfiltration of a GFP-CP fusion displays most of the fluorescence inside the nucleus and within the nucleolus in particular. Nuclear localization of the CP was also confirmed through Western blot analysis of purified nuclei. The significance of several unusual properties of OuMV for replication, virion assembly, and movement is discussed in relation to other positive-strand RNA viruses.
Figures





References
-
- Accotto G. P., Milne R. G. 2008. Ourmiavirus, p. 500–501In Mahy B. W., Van Regenmortel M. H. V.(ed.), Encyclopedia of virology. Elsevier, London, United Kingdom
-
- Agranovsky A. A., et al. 1998. Beet yellows closterovirus HSP70-like protein mediates the cell-to-cell movement of a potexvirus transport-deficient mutant and a hordeivirus-based chimeric virus. J. Gen. Virol. 79:889–895 - PubMed
-
- Aiton M. M., Lennon A. M., Roberts I. M., Harrison B. D. 1988. Two new cassava viruses from Africa, p. 43.In Proceedings of the 5th International Congress of Plant Pathology, Kyoto, Japan Phytopathological Society of Japan, Tokyo, Japan
-
- Annamalai P., Rao A. L. N. 2005. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology 338:96–111 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

