Activities of ICP0 involved in the reversal of silencing of quiescent herpes simplex virus 1
- PMID: 21411540
- PMCID: PMC3126212
- DOI: 10.1128/JVI.02265-10
Activities of ICP0 involved in the reversal of silencing of quiescent herpes simplex virus 1
Abstract
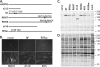
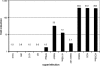
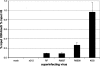
ICP0 is a transcriptional activating protein required for the efficient replication and reactivation of latent herpes simplex virus 1 (HSV-1). Multiple regions of ICP0 contribute its activity, the most prominent of which appears to be the RING finger, which confers E3 ubiquitin ligase activity. A region in the C terminus of ICP0 has also been implicated in several activities, including the disruption of a cellular repressor complex, REST/CoREST/HDAC1/2/LSD1. We used quiescent infection of MRC-5 cells with a virus that does not express immediate-early proteins, followed by superinfection with various viral mutants to quantify the ability of ICP0 variants to reactivate gene expression and alter chromatin structure. Superinfection with wild-type virus resulted in a 400-fold increase in expression from the previously quiescent d109 genome, the removal of heterochromatin and histones from the viral genome, and an increase in histone marks associated with activated transcription. RING finger mutants were unable to reactivate transcription or remove heterochromatin from d109, while mutants that are unable to bind CoREST activate gene expression from quiescent d109, albeit to a lesser degree than the wild-type virus. One such mutant, R8507, resulted in the partial removal of heterochromatin. Infection with R8507 did not result in the hyperacetylation of H3 and H4. The results demonstrate that (i) consistent with previous findings, the RING finger domain of ICP0 is required for the activation of quiescent genomes, (ii) the RF domain is also crucial for the ultimate removal of repressive chromatin, (iii) activities or interactions specified by the carboxy-terminal region of ICP0 significantly contribute to activation, and (iv) while the effects of the R8507 on chromatin are consistent with a role for REST/CoREST/HDAC1/2/LSD1 in the repression of quiescent genomes, the mutation may also affect other activities involved in derepression.
Figures



References
-
- Bannister A. J., et al. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120–124 - PubMed
-
- Barlow P. N., Luisi B., Milner A., Elliott M., Everett R. 1994. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J. Mol. Biol. 237:201–211 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

