Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3
- PMID: 21412236
- PMCID: PMC3077455
- DOI: 10.1038/nature09854
Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3
Abstract
The centromere is a unique chromosomal locus that ensures accurate segregation of chromosomes during cell division by directing the assembly of a multiprotein complex, the kinetochore. The centromere is marked by a conserved variant of conventional histone H3 termed CenH3 or CENP-A (ref. 2). A conserved motif of CenH3, the CATD, defined by loop 1 and helix 2 of the histone fold, is necessary and sufficient for specifying centromere functions of CenH3 (refs 3, 4). The structural basis of this specification is of particular interest. Yeast Scm3 and human HJURP are conserved non-histone proteins that interact physically with the (CenH3-H4)(2) heterotetramer and are required for the deposition of CenH3 at centromeres in vivo. Here we have elucidated the structural basis for recognition of budding yeast (Saccharomyces cerevisiae) CenH3 (called Cse4) by Scm3. We solved the structure of the Cse4-binding domain (CBD) of Scm3 in complex with Cse4 and H4 in a single chain model. An α-helix and an irregular loop at the conserved amino terminus and a shorter α-helix at the carboxy terminus of Scm3(CBD) wraps around the Cse4-H4 dimer. Four Cse4-specific residues in the N-terminal region of helix 2 are sufficient for specific recognition by conserved and functionally important residues in the N-terminal helix of Scm3 through formation of a hydrophobic cluster. Scm3(CBD) induces major conformational changes and sterically occludes DNA-binding sites in the structure of Cse4 and H4. These findings have implications for the assembly and architecture of the centromeric nucleosome.
Figures



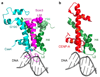
References
-
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and Kinetochores: From epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. - PubMed
-
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. - PubMed
-
- Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. - PubMed
-
- Black BE, et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell. 2007;25:309–322. - PubMed
-
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

