Microbial community composition in sediments resists perturbation by nutrient enrichment
- PMID: 21412346
- PMCID: PMC3160680
- DOI: 10.1038/ismej.2011.22
Microbial community composition in sediments resists perturbation by nutrient enrichment
Abstract
Functional redundancy in bacterial communities is expected to allow microbial assemblages to survive perturbation by allowing continuity in function despite compositional changes in communities. Recent evidence suggests, however, that microbial communities change both composition and function as a result of disturbance. We present evidence for a third response: resistance. We examined microbial community response to perturbation caused by nutrient enrichment in salt marsh sediments using deep pyrosequencing of 16S rRNA and functional gene microarrays targeting the nirS gene. Composition of the microbial community, as demonstrated by both genes, was unaffected by significant variations in external nutrient supply in our sampling locations, despite demonstrable and diverse nutrient-induced changes in many aspects of marsh ecology. The lack of response to external forcing demonstrates a remarkable uncoupling between microbial composition and ecosystem-level biogeochemical processes and suggests that sediment microbial communities are able to resist some forms of perturbation.
Figures

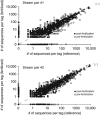

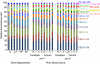
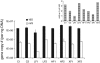
References
-
- Bowen JL, Crump BC, Deegan LA, Hobbie JE. Increased supply of ambient nitrogen has minimal effect on salt marsh bacterial production. Limnol Oceanogr. 2009;54:713–722.

