A combined nucleic acid and protein analysis in Friedreich ataxia: implications for diagnosis, pathogenesis and clinical trial design
- PMID: 21412413
- PMCID: PMC3055871
- DOI: 10.1371/journal.pone.0017627
A combined nucleic acid and protein analysis in Friedreich ataxia: implications for diagnosis, pathogenesis and clinical trial design
Abstract
Background: Friedreich's ataxia (FRDA) is the most common hereditary ataxia among caucasians. The molecular defect in FRDA is the trinucleotide GAA expansion in the first intron of the FXN gene, which encodes frataxin. No studies have yet reported frataxin protein and mRNA levels in a large cohort of FRDA patients, carriers and controls.
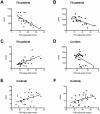
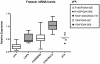
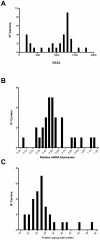
Methodology/principal findings: We enrolled 24 patients with classic FRDA phenotype (cFA), 6 late onset FRDA (LOFA), all homozygous for GAA expansion, 5 pFA cases who harbored the GAA expansion in compound heterozygosis with FXN point mutations (namely, p.I154F, c.482+3delA, p.R165P), 33 healthy expansion carriers, and 29 healthy controls. DNA was genotyped for GAA expansion, mRNA/FXN was quantified in real-time, and frataxin protein was measured using lateral-flow immunoassay in peripheral blood mononuclear cells (PBMCs). Mean residual levels of frataxin, compared to controls, were 35.8%, 65.6%, 33%, and 68.7% in cFA, LOFA, pFA and healthy carriers, respectively. Comparison of both cFA and pFA with controls resulted in 100% sensitivity and specificity, but there was overlap between LOFA, carriers and controls. Frataxin levels correlated inversely with GAA1 and GAA2 expansions, and directly with age at onset. Messenger RNA expression was reduced to 19.4% in cFA, 50.4% in LOFA, 52.7% in pFA, 53.0% in carriers, as compared to controls (p<0.0001). mRNA levels proved to be diagnostic when comparing cFA with controls resulting in 100% sensitivity and specificity. In cFA and LOFA patients mRNA levels correlated directly with protein levels and age at onset, and inversely with GAA1 and GAA2.
Conclusion/significance: We report the first explorative study on combined frataxin and mRNA levels in PBMCs from a cohort of FRDA patients, carriers and healthy individuals. Lateral-flow immunoassay differentiated cFA and pFA patients from controls, whereas determination of mRNA in q-PCR was sensitive and specific only in cFA.
Conflict of interest statement
Figures


References
-
- Harding AE. Friedreich's ataxia: a clinical and genetic stud of 90 families with analysis of early diagnostic criteria and intrafamilial clustering of clinical features. Brain. 1981;104:589–620. - PubMed
-
- Filla A, De Michele G, Coppola G, Federico A, Vita G, et al. Accuracy of clinical diagnostic criteria for Friedreich's ataxia. Mov Disord. 2000;15:1255–1258. - PubMed
-
- De Michele G, Perrone F, Filla A, Mirante E, Giordano M, et al. Age of onset, sex, and cardiomyopathy as predictors of disability and survival in Friedreich's disease: a retrospective study on 119 patients. Neurology; 1996;47:1260–4. - PubMed
-
- Campuzano V, Montermini L, Moltò MD, Pianese L, Cossée M, et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

