Drop-on-demand single cell isolation and total RNA analysis
- PMID: 21412416
- PMCID: PMC3055874
- DOI: 10.1371/journal.pone.0017455
Drop-on-demand single cell isolation and total RNA analysis
Abstract
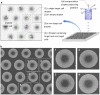
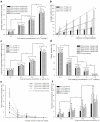
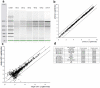
Technologies that rapidly isolate viable single cells from heterogeneous solutions have significantly contributed to the field of medical genomics. Challenges remain both to enable efficient extraction, isolation and patterning of single cells from heterogeneous solutions as well as to keep them alive during the process due to a limited degree of control over single cell manipulation. Here, we present a microdroplet based method to isolate and pattern single cells from heterogeneous cell suspensions (10% target cell mixture), preserve viability of the extracted cells (97.0±0.8%), and obtain genomic information from isolated cells compared to the non-patterned controls. The cell encapsulation process is both experimentally and theoretically analyzed. Using the isolated cells, we identified 11 stem cell markers among 1000 genes and compare to the controls. This automated platform enabling high-throughput cell manipulation for subsequent genomic analysis employs fewer handling steps compared to existing methods.
Conflict of interest statement
Figures

References
-
- Bianco P, Robey PG. Stem cells in tissue engineering. Nature. 2001;414:118–121. - PubMed
-
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, et al. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell. 2007;131:861–872. - PubMed
-
- Zipori D. The Stem State: Plasticity Is Essential, Whereas Self-Renewal and Hierarchy Are Optional. Stem Cells. 2005;23:719–726. - PubMed
-
- Slack JMW. Origin of Stem Cells in Organogenesis. Science. 2008;322:1498–1501. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

