Identification of intragenic deletions and duplication in the FLCN gene in Birt-Hogg-Dubé syndrome
- PMID: 21412933
- PMCID: PMC3075348
- DOI: 10.1002/gcc.20872
Identification of intragenic deletions and duplication in the FLCN gene in Birt-Hogg-Dubé syndrome
Abstract
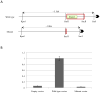
Birt-Hogg-Dubé syndrome (BHDS), caused by germline mutations in the folliculin (FLCN) gene, predisposes individuals to develop fibrofolliculomas, pulmonary cysts, spontaneous pneumothoraces, and kidney cancer. The FLCN mutation detection rate by bidirectional DNA sequencing in the National Cancer Institute BHDS cohort was 88%. To determine if germline FLCN intragenic deletions/duplications were responsible for BHDS in families lacking FLCN sequence alterations, 23 individuals from 15 unrelated families with clinically confirmed BHDS but no sequence variations were analyzed by real-time quantitative PCR (RQ-PCR) using primers for all 14 exons. Multiplex ligation-dependent probe amplification (MLPA) assay and array-based comparative genomic hybridization (aCGH) were utilized to confirm and fine map the rearrangements. Long-range PCR followed by DNA sequencing was used to define the breakpoints. We identified six unique intragenic deletions in nine patients from six different BHDS families including four involving exon 1, one that spanned exons 2-5, and one that encompassed exons 7-14 of FLCN. Four of the six deletion breakpoints were mapped, revealing deletions ranging from 5688 to 9189 bp. In addition, one 1341 bp duplication, which included exons 10 and 11, was identified and mapped. This report confirms that large intragenic FLCN deletions can cause BHDS and documents the first large intragenic FLCN duplication in a BHDS patient. Additionally, we identified a deletion "hot spot" in the 5'-noncoding-exon 1 region that contains the putative FLCN promoter based on a luciferase reporter assay. RQ-PCR, MLPA and aCGH may be used for clinical molecular diagnosis of BHDS in patients who are FLCN mutation-negative by DNA sequencing.
Copyright © 2011 Wiley-Liss, Inc.
Conflict of interest statement
None.
Figures




References
-
- Ahvenainen T, Lehtonen HJ, Lehtonen R, Vahteristo P, Aittomaki K, Baynam G, Dommering C, Eng C, Gruber SB, Gronberg H, Harvima R, Herva R, Hietala M, Kujala M, Kaariainen H, Sunde L, Vierimaa O, Pollard PJ, Tomlinson IP, Bjorck E, Aaltonen LA, Launonen V. Mutation screening of fumarate hydratase by multiplex ligation-dependent probe amplification: detection of exonic deletion in a patient with leiomyomatosis and renal cell cancer. Cancer Genet Cytogenet. 2008;183:83–88. - PubMed
-
- Baba M, Hong SB, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, Esposito D, Gillette WK, Hopkins RF, III, Hartley JL, Furihata M, Oishi S, Zhen W, Burke TR, Jr, Linehan WM, Schmidt LS, Zbar B. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci U S A. 2006;103:15552–15557. - PMC - PubMed
-
- Birt AR, Hogg GR, Dube WJ. Hereditary multiple fibrofolliculomas with trichodiscomas and acrochordons. Arch Dermatol. 1977;113:1674–1677. - PubMed
-
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. - PubMed
-
- De La Torre C, Ocampo C, Doval IG, Losada A, Cruces MJ. Acrochordons are not a component of the Birt-Hogg-Dube syndrome: does this syndrome exist? Case reports and review of the literature. Am J Dermatopathol. 1999;21:369–374. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

