Generation of mice with a conditional allele for the p75(NTR) neurotrophin receptor gene
- PMID: 21413144
- PMCID: PMC3543998
- DOI: 10.1002/dvg.20747
Generation of mice with a conditional allele for the p75(NTR) neurotrophin receptor gene
Abstract
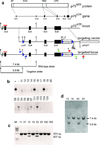
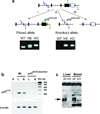
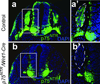
The p75(NTR) neurotrophin receptor has been implicated in multiple biological and pathological processes. While significant advances have recently been made in understanding the physiologic role of p75(NTR) , many details and aspects remain to be determined. This is in part because the two existing knockout mouse models (Exons 3 or 4 deleted, respectively), both display features that defy definitive conclusions. Here we describe the generation of mice that carry a conditional p75(NTR) (p75(NTR-FX) ) allele made by flanking Exons 4-6, which encode the transmembrane and all cytoplasmic domains, by loxP sites. To validate this novel conditional allele, both neural crest-specific p75(NTR) /Wnt1-Cre mutants and conventional p75(NTR) null mutants were generated. Both mutants displayed abnormal hind limb reflexes, implying that loss of p75(NTR) in neural crest-derived cells causes a peripheral neuropathy similar to that seen in conventional p75(NTR) mutants. This novel conditional p75(NTR) allele will offer new opportunities to investigate the role of p75(NTR) in specific tissues and cells.
Copyright © 2011 Wiley-Periodicals, Inc.
Figures


Similar articles
-
Schwann cell p75NTR prevents spontaneous sensory reinnervation of the adult spinal cord.Brain. 2010 Feb;133(Pt 2):421-32. doi: 10.1093/brain/awp316. Epub 2010 Jan 3. Brain. 2010. PMID: 20047901
-
Expression of Ret-, p75(NTR)-, Phox2a-, Phox2b-, and tyrosine hydroxylase-immunoreactivity by undifferentiated neural crest-derived cells and different classes of enteric neurons in the embryonic mouse gut.Dev Dyn. 1999 Oct;216(2):137-52. doi: 10.1002/(SICI)1097-0177(199910)216:2<137::AID-DVDY5>3.0.CO;2-6. Dev Dyn. 1999. PMID: 10536054
-
Death Domain Signaling by Disulfide-Linked Dimers of the p75 Neurotrophin Receptor Mediates Neuronal Death in the CNS.J Neurosci. 2016 May 18;36(20):5587-95. doi: 10.1523/JNEUROSCI.4536-15.2016. J Neurosci. 2016. PMID: 27194337 Free PMC article.
-
Role of p75 neurotrophin receptor in stem cell biology: more than just a marker.Cell Mol Life Sci. 2014 Jul;71(13):2467-81. doi: 10.1007/s00018-014-1564-9. Epub 2014 Jan 31. Cell Mol Life Sci. 2014. PMID: 24481864 Free PMC article. Review.
-
The Role of Neurotrophin Signaling in Gliomagenesis: A Focus on the p75 Neurotrophin Receptor (p75NTR/CD271).Vitam Horm. 2017;104:367-404. doi: 10.1016/bs.vh.2016.11.001. Epub 2017 Jan 4. Vitam Horm. 2017. PMID: 28215302 Review.
Cited by
-
Role of proNGF/p75 signaling in bladder dysfunction after spinal cord injury.J Clin Invest. 2018 May 1;128(5):1772-1786. doi: 10.1172/JCI97837. Epub 2018 Mar 26. J Clin Invest. 2018. PMID: 29584618 Free PMC article.
-
Excess cerebellar granule neurons induced by the absence of p75NTR during development elicit social behavior deficits in mice.Front Mol Neurosci. 2023 May 25;16:1147597. doi: 10.3389/fnmol.2023.1147597. eCollection 2023. Front Mol Neurosci. 2023. PMID: 37305555 Free PMC article.
-
TAp73 knockout mice show morphological and functional nervous system defects associated with loss of p75 neurotrophin receptor.Proc Natl Acad Sci U S A. 2013 Nov 19;110(47):18952-7. doi: 10.1073/pnas.1221172110. Epub 2013 Nov 4. Proc Natl Acad Sci U S A. 2013. PMID: 24190996 Free PMC article.
-
The p75NTR Influences Cerebellar Circuit Development and Adult Behavior via Regulation of Cell Cycle Duration of Granule Cell Progenitors.J Neurosci. 2019 Nov 13;39(46):9119-9129. doi: 10.1523/JNEUROSCI.0990-19.2019. Epub 2019 Oct 3. J Neurosci. 2019. PMID: 31582529 Free PMC article.
-
Cerebellar Purkinje cell p75 neurotrophin receptor and autistic behavior.Transl Psychiatry. 2014 Jul 29;4(7):e416. doi: 10.1038/tp.2014.55. Transl Psychiatry. 2014. PMID: 25072321 Free PMC article.
References
-
- Barker PA. p75NTR is positively promiscuous: novel partners and new insights. Neuron. 2004;42:529–533. - PubMed
-
- Bronfman FC. Metalloproteases and gamma-secretase: new membrane partners regulating p75 neurotrophin receptor signaling? J Neurochem. 2007;103(Suppl 1):91–100. - PubMed
-
- Chai Y, Jiang X, Ito Y, Bringas P, Jr, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. - PubMed
-
- Dudas M, Sridurongrit S, Nagy A, Okazaki K, Kaartinen V. Craniofacial defects in mice lacking BMP type I receptor Alk2 in neural crest cells. Mech.Dev. 2004;121:173–182. - PubMed
-
- Harlow E, Lane D. A Laboratory Manual. CSH Press; 1988. Antibodies.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

