Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase ε
- PMID: 21414850
- PMCID: PMC3652408
- DOI: 10.1016/j.dnarep.2011.02.001
Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase ε
Abstract
During DNA synthesis in vitro using dNTP and rNTP concentrations present in vivo, yeast replicative DNA polymerases α, δ and ɛ (Pols α, δ and ɛ) stably incorporate rNTPs into DNA. rNTPs are also incorporated during replication in vivo, and they are repaired in an RNase H2-dependent manner. In strains encoding a mutator allele of Pol ɛ (pol2-M644G), failure to remove rNMPs from DNA due to deletion of the RNH201 gene encoding the catalytic subunit of RNase H2, results in deletion of 2-5 base pairs in short repetitive sequences. Deletion rates depend on the orientation of the reporter gene relative to a nearby replication origin, suggesting that mutations result from rNMPs incorporated during replication. Here we demonstrate that 2-5 base pair deletion mutagenesis also strongly increases in rnh201Δ strains encoding wild type DNA polymerases. As in the pol2-M644G strains, the deletions occur at repetitive sequences and are orientation-dependent, suggesting that mismatches involving misaligned strands arise that could be subject to mismatch repair. Unexpectedly however, 2-5 base pair deletion rates resulting from loss of RNH201 in the pol2-M644G strain are unaffected by concomitant loss of MSH3, MSH6, or both. It could be that the mismatch repair machinery is unable to repair mismatches resulting from unrepaired rNMPs incorporated into DNA by M644G Pol ɛ, but this possibility is belied by the observation that Msh2-Msh6 can bind to a ribonucleotide-containing mismatch. Alternatively, following incorporation of rNMPs by M644G Pol ɛ during replication, the conversion of unrepaired rNMPs into mutations may occur outside the context of replication, e.g., during the repair of nicks resulting from rNMPs in DNA. The results make interesting predictions that can be tested.
Published by Elsevier B.V.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
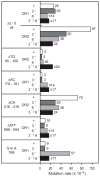

 ), POL2 RNH201 URA3 in orientation 2 (
), POL2 RNH201 URA3 in orientation 2 (
 ), POL2 rnh201Δ URA3 in orientation 2 (■). (B). Results for pol2-M644G strains that are either RNH201 (+) or rnh201Δ (−). Individual rates, above the columns, were calculated as in (A) from results in Table 1 and Figure 4 in [7]. Columns are shaded for pol2-M644G RNH201 URA3 in orientation 1 (□), pol2-M644G rnh201Δ URA3 in orientation 1 (
), POL2 rnh201Δ URA3 in orientation 2 (■). (B). Results for pol2-M644G strains that are either RNH201 (+) or rnh201Δ (−). Individual rates, above the columns, were calculated as in (A) from results in Table 1 and Figure 4 in [7]. Columns are shaded for pol2-M644G RNH201 URA3 in orientation 1 (□), pol2-M644G rnh201Δ URA3 in orientation 1 (
 ), pol2-M644G RNH201 URA3 in orientation 2 (
), pol2-M644G RNH201 URA3 in orientation 2 (
 ), pol2-M644G rnh201Δ URA3 in orientation 2 (■).
), pol2-M644G rnh201Δ URA3 in orientation 2 (■).
 ), pol2-M644G rnh201Δ msh6Δ (
), pol2-M644G rnh201Δ msh6Δ (
 ), pol2-M644G rnh201Δ msh3Δ msh6Δ (■). Orientation of URA3 is designated as OR1 or OR2.
), pol2-M644G rnh201Δ msh3Δ msh6Δ (■). Orientation of URA3 is designated as OR1 or OR2.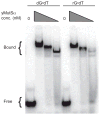
References
-
- Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

