Protein misfolding and oxidative stress promote glial-mediated neurodegeneration in an Alexander disease model
- PMID: 21414908
- PMCID: PMC3082397
- DOI: 10.1523/JNEUROSCI.3410-10.2011
Protein misfolding and oxidative stress promote glial-mediated neurodegeneration in an Alexander disease model
Abstract
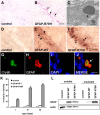
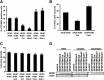
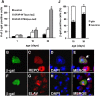
Although alterations in glial structure and function commonly accompany death of neurons in neurodegenerative diseases, the role glia play in modulating neuronal loss is poorly understood. We have created a model of Alexander disease in Drosophila by expressing disease-linked mutant versions of glial fibrillary acidic protein (GFAP) in fly glia. We find aggregation of mutant human GFAP into inclusions bearing the hallmarks of authentic Rosenthal fibers. We also observe significant toxicity of mutant human GFAP to glia, which is mediated by protein aggregation and oxidative stress. Both protein aggregation and oxidative stress contribute to activation of a robust autophagic response in glia. Toxicity of mutant GFAP to glial cells induces a non-cell-autonomous stress response and subsequent apoptosis in neurons, which is dependent on glial glutamate transport. Our findings thus establish a simple genetic model of Alexander disease and further identify cellular pathways critical for glial-induced neurodegeneration.
Figures






References
-
- Arrigo AP, Simon S, Gibert B, Kretz-Remy C, Nivon M, Czekalla A, Guillet D, Moulin M, Diaz-Latoud C, Vicart P. Hsp27 (HspB1) and alphaB-crystallin (HspB5) as therapeutic targets. FEBS Lett. 2007;581:3665–3674. - PubMed
-
- Azad MB, Chen Y, Gibson SB. Regulation of autophagy by reactive oxygen species (ROS): implications for cancer progression and treatment. Antioxid Redox Signal. 2009;11:777–790. - PubMed
-
- Benzer S. From the gene to behavior. JAMA. 1971;218:1015–1022. - PubMed
-
- Bonini NM, Fortini ME. Human neurodegenerative disease modeling using Drosophila. Annu Rev Neurosci. 2003;26:627–656. - PubMed
-
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
