Second-order selection for evolvability in a large Escherichia coli population
- PMID: 21415350
- PMCID: PMC3176658
- DOI: 10.1126/science.1198914
Second-order selection for evolvability in a large Escherichia coli population
Abstract
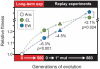
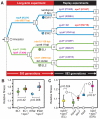
In theory, competition between asexual lineages can lead to second-order selection for greater evolutionary potential. To test this hypothesis, we revived a frozen population of Escherichia coli from a long-term evolution experiment and compared the fitness and ultimate fates of four genetically distinct clones. Surprisingly, two clones with beneficial mutations that would eventually take over the population had significantly lower competitive fitness than two clones with mutations that later went extinct. By replaying evolution many times from these clones, we showed that the eventual winners likely prevailed because they had greater potential for further adaptation. Genetic interactions that reduce the benefit of certain regulatory mutations in the eventual losers appear to explain, at least in part, why they were outcompeted.
Figures


References
-
- Pigliucci M. Is evolvability evolvable? Nat. Rev. Genet. 2008;9:75. - PubMed
-
- Weinreich DM, Watson RA, Chao L. Perspective: Sign epistasis and genetic constraint on evolutionary trajectories. Evolution. 2005;59:1165. - PubMed
-
- Chao L, Cox EC. Competition between high and low mutating strains of Escherichia coli. Evolution. 1983;37:125. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

