Thin filament mutations: developing an integrative approach to a complex disorder
- PMID: 21415410
- PMCID: PMC3075069
- DOI: 10.1161/CIRCRESAHA.110.224170
Thin filament mutations: developing an integrative approach to a complex disorder
Abstract
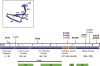
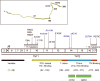
Sixteen years ago, mutations in cardiac troponin (Tn)T and α-tropomyosin were linked to familial hypertrophic cardiomyopathy, thus transforming the disorder from a disease of the β-myosin heavy chain to a disease of the cardiac sarcomere. From the outset, studies suggested that mutations in the regulatory thin filament caused a complex, heterogeneous pattern of ventricular remodeling with wide variations in clinical expression. To date, the clinical heterogeneity is well matched by an extensive array of nearly 100 independent mutations in all components of the cardiac thin filament. Significant advances in our understanding of the biophysics of myofilament activation, coupled to the emerging evidence that thin filament linked cardiomyopathies are progressive, suggests that a renewed focus on the most proximal events in both the molecular and clinical pathogenesis of the disease will be necessary to achieve the central goal of using genotype information to manage affected patients. In this review, we examine the existing biophysical and clinical evidence in support of a more proximal definition of thin filament cardiomyopathies. In addition, new high-resolution, integrated approaches are presented to help define the way forward as the field works toward developing a more robust link between genotype and phenotype in this complex disorder.
Figures




Similar articles
-
Rescue of tropomyosin-induced familial hypertrophic cardiomyopathy mice by transgenesis.Am J Physiol Heart Circ Physiol. 2007 Aug;293(2):H949-58. doi: 10.1152/ajpheart.01341.2006. Epub 2007 Apr 6. Am J Physiol Heart Circ Physiol. 2007. PMID: 17416600
-
Sarcomeric proteins and familial hypertrophic cardiomyopathy: linking mutations in structural proteins to complex cardiovascular phenotypes.Heart Fail Rev. 2005 Sep;10(3):237-48. doi: 10.1007/s10741-005-5253-5. Heart Fail Rev. 2005. PMID: 16416046 Review.
-
Clinically Divergent Mutation Effects on the Structure and Function of the Human Cardiac Tropomyosin Overlap.Biochemistry. 2017 Jul 5;56(26):3403-3413. doi: 10.1021/acs.biochem.7b00266. Epub 2017 Jun 21. Biochemistry. 2017. PMID: 28603979 Free PMC article.
-
Microvascular function is selectively impaired in patients with hypertrophic cardiomyopathy and sarcomere myofilament gene mutations.J Am Coll Cardiol. 2011 Aug 16;58(8):839-48. doi: 10.1016/j.jacc.2011.05.018. J Am Coll Cardiol. 2011. PMID: 21835320
-
Alpha-tropomyosin mutations in inherited cardiomyopathies.J Muscle Res Cell Motil. 2013 Aug;34(3-4):285-94. doi: 10.1007/s10974-013-9358-5. Epub 2013 Sep 5. J Muscle Res Cell Motil. 2013. PMID: 24005378 Review.
Cited by
-
A study of tropomyosin's role in cardiac function and disease using thin-filament reconstituted myocardium.J Muscle Res Cell Motil. 2013 Aug;34(3-4):295-310. doi: 10.1007/s10974-013-9343-z. Epub 2013 May 23. J Muscle Res Cell Motil. 2013. PMID: 23700264 Free PMC article. Review.
-
p90 ribosomal S6 kinase 3 contributes to cardiac insufficiency in α-tropomyosin Glu180Gly transgenic mice.Am J Physiol Heart Circ Physiol. 2013 Oct 1;305(7):H1010-9. doi: 10.1152/ajpheart.00237.2013. Epub 2013 Aug 2. Am J Physiol Heart Circ Physiol. 2013. PMID: 23913705 Free PMC article.
-
Docking Troponin T onto the Tropomyosin Overlapping Domain of Thin Filaments.Biophys J. 2020 Jan 21;118(2):325-336. doi: 10.1016/j.bpj.2019.11.3393. Epub 2019 Dec 6. Biophys J. 2020. PMID: 31864661 Free PMC article.
-
The genetics of hypertrophic cardiomyopathy.Glob Cardiol Sci Pract. 2018 Aug 12;2018(3):36. doi: 10.21542/gcsp.2018.36. Glob Cardiol Sci Pract. 2018. PMID: 30393648 Free PMC article. Review.
-
Molecule specific effects of PKA-mediated phosphorylation on rat isolated heart and cardiac myofibrillar function.Arch Biochem Biophys. 2016 Jul 1;601:22-31. doi: 10.1016/j.abb.2016.01.019. Epub 2016 Feb 15. Arch Biochem Biophys. 2016. PMID: 26854722 Free PMC article.
References
-
- Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, Seidman JG. A molecular basis for familial hypertrophic cardiomyopathy: A beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. - PubMed
-
- Thierfelder L, Watkins H, MacRae C, Lamas R, McKenna W, Vosberg HP, Seidman JG, Seidman CE. Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: A disease of the sarcomere. Cell. 1994;77:701–712. - PubMed
-
- Kimura A, Harada H, Park JE, Nishi H, Satoh M, Takahashi M, Hiroi S, Sasaoka T, Ohbuchi N, Nakamura T, Koyanagi T, Hwang TH, Choo JA, Chung KS, Hasegawa A, Nagai R, Okazaki O, Nakamura H, Matsuzaki M, Sakamoto T, Toshima H, Koga Y, Imaizumi T, Sasazuki T. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nature. 1997;16:379–382. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

