The importance of epigenetic alterations in the development of epstein-barr virus-related lymphomas
- PMID: 21416002
- PMCID: PMC3033174
- DOI: 10.4084/MJHID.2009.012
The importance of epigenetic alterations in the development of epstein-barr virus-related lymphomas
Abstract
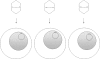
Epstein-Barr virus (EBV), a human gammaherpesvirus, is associated with a series of malignant tumors. These include lymphomas (Burkitt's lymphoma, Hodgkin's disease, T/NK-cell lymphoma, post-transplant lymphoproliferative disease, AIDS-associated lymphoma, X-linked lymphoproliferative syndrome), carcinomas (nasopharyngeal carcinoma, gastric carcinoma, carcinomas of major salivary glands, thymic carcinoma, mammary carcinoma) and a sarcoma (leiomyosarcoma). The latent EBV genomes persist in the tumor cells as circular episomes, co-replicating with the cellular DNA once per cell cycle. The expression of latent EBV genes is cell type specific due to the strict epigenetic control of their promoters. DNA methylation, histone modifications and binding of key cellular regulatory proteins contribute to the regulation of alternative promoters for transcripts encoding the nuclear antigens EBNA1 to 6 and affect the activity of promoters for transcripts encoding transmembrane proteins (LMP1, LMP2A, LMP2B). In addition to genes transcribed by RNA polymerase II, there are also two RNA polymerase III transcribed genes in the EBV genome (EBER 1 and 2). The 5' and internal regulatory sequences of EBER 1 and 2 transcription units are invariably unmethylated. The highly abundant EBER 1 and 2 RNAs are not translated to protein. Based on the cell type specific epigenetic marks associated with latent EBV genomes one can distinguish between viral epigenotypes that differ in transcriptional activity in spite of having an identical (or nearly identical) DNA sequence. Whereas latent EBV genomes are regularly targeted by epigenetic control mechanisms in different cell types, EBV encoded proteins may, in turn, affect the activity of a set of cellular promoters by interacting with the very same epigenetic regulatory machinery. There are EBNA1 binding sites in the human genome. Because high affinity binding of EBNA1 to its recognition sites is known to specify sites of DNA demethylation, we suggest that binding of EBNA1 to its cellular target sites may elicit local demethylation and contribute thereby to the activation of silent cellular promoters. EBNA2 interacts with histone acetyltransferases, and EBNALP (EBNA5) coactivates transcription by displacing histone deacetylase 4 from EBNA2-bound promoter sites. EBNA3C (EBNA6) seems to be associated both with histone acetylases and deacetylases, although in separate complexes. LMP1, a transmembrane protein involved in malignant transformation, can affect both alternative systems of epigenetic memory, DNA methylation and the Polycomb-trithorax group of protein complexes. In epithelial cells LMP1 can up-regulate DNA methyltransferases and, in Hodgkin lymphoma cells, induce the Polycomb group protein Bmi-1. In addition, LMP1 can also modulate cellular gene expression programs by affecting, via the NF-κB pathway, levels of cellular microRNAs miR-146a and miR-155. These interactions may result in epigenetic dysregulation and subsequent cellular dysfunctions that may manifest in or contribute to the development of pathological changes (e.g. initiation and progression of malignant neoplasms, autoimmune phenomena, immunodeficiency). Thus, Epstein-Barr virus, similarly to other viruses and certain bacteria, may induce pathological changes by epigenetic reprogramming of host cells. Elucidation of the epigenetic consequences of EBV-host interactions (within the framework of the emerging new field of patho-epigenetics) may have important implications for therapy and disease prevention, because epigenetic processes are reversible and continuous silencing of EBV genes contributing to patho-epigenetic changes may prevent disease development.
Figures
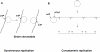
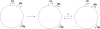
Similar articles
-
Epigenetic regulation of latent Epstein-Barr virus promoters.Biochim Biophys Acta. 2010 Mar-Apr;1799(3-4):228-35. doi: 10.1016/j.bbagrm.2009.10.005. Epub 2009 Oct 22. Biochim Biophys Acta. 2010. PMID: 19853674 Review.
-
Host cell-dependent expression of latent Epstein-Barr virus genomes: regulation by DNA methylation.Adv Cancer Res. 2003;89:133-56. doi: 10.1016/s0065-230x(03)01004-2. Adv Cancer Res. 2003. PMID: 14587872 Review.
-
Epigenotypes of latent herpesvirus genomes.Curr Top Microbiol Immunol. 2006;310:61-80. doi: 10.1007/3-540-31181-5_5. Curr Top Microbiol Immunol. 2006. PMID: 16909907 Review.
-
The DNA loop release factor WAPL suppresses Epstein-Barr virus latent membrane protein expression to maintain the highly restricted latency I program.bioRxiv [Preprint]. 2024 May 9:2024.05.09.593401. doi: 10.1101/2024.05.09.593401. bioRxiv. 2024. Update in: PLoS Pathog. 2024 Sep 6;20(9):e1012525. doi: 10.1371/journal.ppat.1012525. PMID: 38766209 Free PMC article. Updated. Preprint.
-
Epstein-Barr Virus Nuclear Antigen Leader Protein Coactivates EP300.J Virol. 2018 Apr 13;92(9):e02155-17. doi: 10.1128/JVI.02155-17. Print 2018 May 1. J Virol. 2018. PMID: 29467311 Free PMC article.
Cited by
-
Proteoglycan expression correlates with the phenotype of malignant and non-malignant EBV-positive B-cell lines.Oncotarget. 2015 Dec 22;6(41):43529-39. doi: 10.18632/oncotarget.5984. Oncotarget. 2015. PMID: 26527314 Free PMC article.
-
Network Pharmacology to Uncover the Molecular Mechanisms of Action of LeiGongTeng for the Treatment of Nasopharyngeal Carcinoma.Med Sci Monit Basic Res. 2020 May 25;26:e923431. doi: 10.12659/MSMBR.923431. Med Sci Monit Basic Res. 2020. PMID: 32448862 Free PMC article.
-
EBV Chronic Infections.Mediterr J Hematol Infect Dis. 2010 Aug 10;2(1):e2010022. doi: 10.4084/MJHID.2010.022. Mediterr J Hematol Infect Dis. 2010. PMID: 21415952 Free PMC article.
-
Role of immune escape mechanisms in Hodgkin's lymphoma development and progression: a whole new world with therapeutic implications.Clin Dev Immunol. 2012;2012:756353. doi: 10.1155/2012/756353. Epub 2012 Aug 15. Clin Dev Immunol. 2012. PMID: 22927872 Free PMC article. Review.
-
Polymicrobial infection and bacterium-mediated epigenetic modification of DNA tumor viruses contribute to pathogenesis.mBio. 2014 Apr 29;5(3):e01015-14. doi: 10.1128/mBio.01015-14. mBio. 2014. PMID: 24781742 Free PMC article. Review.
References
-
- Niller HH, Wolf H, Minarovits J. Epstein-Barr Virus. In: Minarovits J, Gonczol E, Valyi-Nagy T, editors. Latency Strategies of Herpesviruses. Springer; 2007. pp. 154–191.
-
- Epstein MA, Achong BG, Barr YM. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet. 1964;1:702–703. - PubMed
-
- Li H, Minarovits J. Host cell-dependent expression of latent Epstein-Barr virus genomes: Regulation by DNA methylation. Advances in Cancer Res. 2003;89:133–156. - PubMed
-
- Dillner J, Kallin B. The Epstein-Barr virus proteins. Adv Cancer Res. 1988;50:95–158. - PubMed
LinkOut - more resources
Full Text Sources
