Cytoskeletal protein kinases: titin and its relations in mechanosensing
- PMID: 21416260
- PMCID: PMC3114093
- DOI: 10.1007/s00424-011-0946-1
Cytoskeletal protein kinases: titin and its relations in mechanosensing
Abstract
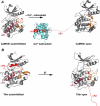
Titin, the giant elastic ruler protein of striated muscle sarcomeres, contains a catalytic kinase domain related to a family of intrasterically regulated protein kinases. The most extensively studied member of this branch of the human kinome is the Ca(2+)-calmodulin (CaM)-regulated myosin light-chain kinases (MLCK). However, not all kinases of the MLCK branch are functional MLCKs, and about half lack a CaM binding site in their C-terminal autoinhibitory tail (AI). A unifying feature is their association with the cytoskeleton, mostly via actin and myosin filaments. Titin kinase, similar to its invertebrate analogue twitchin kinase and likely other "MLCKs", is not Ca(2+)-calmodulin-activated. Recently, local protein unfolding of the C-terminal AI has emerged as a common mechanism in the activation of CaM kinases. Single-molecule data suggested that opening of the TK active site could also be achieved by mechanical unfolding of the AI. Mechanical modulation of catalytic activity might thus allow cytoskeletal signalling proteins to act as mechanosensors, creating feedback mechanisms between cytoskeletal tension and tension generation or cellular remodelling. Similar to other MLCK-like kinases like DRAK2 and DAPK1, TK is linked to protein turnover regulation via the autophagy/lysosomal system, suggesting the MLCK-like kinases have common functions beyond contraction regulation.
Figures






References
-
- Agarkova I, Perriard JC. The M-band: an elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 2005;15:477–85. - PubMed
-
- Agarkova I, Ehler E, Lange S, Schoenauer R, Perriard JC. M-band: a safeguard for sarcomere stability? J Muscle Res Cell Motil. 2003;24:191–203. - PubMed
-
- Akiyama N, Ohnuki Y, Kunioka Y, Saeki Y, Yamada T. Transverse stiffness of myofibrils of skeletal and cardiac muscles studied by atomic force microscopy. J Physiol Sci. 2006;56:145–151. - PubMed
-
- Amodeo P, Castiglione Morelli MA, Strazzullo G, Fucile P, Gautel M, Motta A. Kinase recognition by calmodulin: modeling the interaction with the autoinhibitory region of human cardiac titin kinase. J Mol Biol. 2001;306:81–95. - PubMed
-
- Avrova SV, Shelud'ko NS, Borovikov YS, Galler S. Twitchin of mollusc smooth muscles can induce “catch”-like properties in human skeletal muscle: support for the assumption that the “catch” state involves twitchin linkages between myofilaments. J Comp Physiol B. 2009;179:945–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

