Analysis of in situ manganese(II) oxidation in the Columbia River and offshore plume: linking Aurantimonas and the associated microbial community to an active biogeochemical cycle
- PMID: 21418498
- PMCID: PMC5657485
- DOI: 10.1111/j.1462-2920.2011.02462.x
Analysis of in situ manganese(II) oxidation in the Columbia River and offshore plume: linking Aurantimonas and the associated microbial community to an active biogeochemical cycle
Abstract
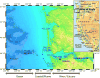
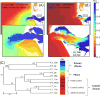
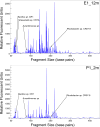
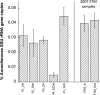
The Columbia River is a major source of dissolved nutrients and trace metals for the west coast of North America. A large proportion of these nutrients are sourced from the Columbia River Estuary, where coastal and terrestrial waters mix and resuspend particulate matter within the water column. As estuarine water is discharged off the coast, it transports the particulate matter, dissolved nutrients and microorganisms forming nutrient-rich and metabolically dynamic plumes. In this study, bacterial manganese oxidation within the plume and estuary was investigated during spring and neap tides. The microbial community proteome was fractionated and assayed for Mn oxidation activity. Proteins from the outer membrane and the loosely bound outer membrane fractions were separated using size exclusion chromatography and Mn(II)-oxidizing eluates were analysed with tandem mass spectrometry to identify potential Mn oxidase protein targets. Multi-copper oxidase (MCO) and haem-peroxidase enzymes were identified in active fractions. T-RFLP profiles and cluster analysis indicates that organisms and bacterial communities capable of oxidizing Mn(II) can be sourced from the Columbia River estuary and nearshore coastal ocean. These organisms are producing up to 10 fM MnO₂ cell⁻¹ day⁻¹. Evidence for the presence of Mn(II)-oxidizing bacterial isolates from the genera Aurantimonas, Rhodobacter, Bacillus and Shewanella was found in T-RFLP profiles. Specific Q-PCR probes were designed to target potential homologues of the Aurantimonas manganese oxidizing peroxidase (Mop). By comparing total Mop homologues, Aurantimonas SSU rRNA and total bacterial SSU rRNA gene copies, it appears that Aurantimonas can only account for ~1.7% of the peroxidase genes quantified. Under the broad assumption that at least some of the peroxidase homologues quantified are involved in manganese oxidation, it is possible that other organisms oxidize manganese via peroxidases.
© 2011 Society for Applied Microbiology and Blackwell Publishing Ltd.
Figures


References
-
- Aguilar-Islas AM, Bruland KW. Dissolved manganese and silicic acid in the Columbia River plume: A major source to the California current and coastal waters off Washington and Oregon. Mar Chem. 2006;101:233–247.
-
- Anderson CR, Dick GJ, Chu M-L, Cho J-C, Davis RE, Bräuer SL, Tebo BM. Aurantimonas manganoxydans, sp. nov. and Aurantimonas litoralis, sp. nov.: Mn(II) oxidizing representatives of a globally distributed clade of alpha-Proteobacteria from the order Rhizobiales. Geomicrobiol J. 2009a;26:189–198. - PMC - PubMed
-
- Barnes CA, Duxbury AC, Morse BA. Circulation and selected properties of the Columbia River effluent at sea. In: Pruter AT, Alverson DL, editors. The Columbia River Estuary and Adjacent Ocean Waters. Seattle: University of Washington Press; 1972. pp. 41–80.
-
- Baross JA, Crump BC, Simenstad CA. Elevated 'microbial loop' activities in the Columbia River estuarine turbidity maximum. In: Dyer KR, Orth RJ, editors. Changes in Fluxes in Estuaries: Implications from Science to Management. Fredensborg, Denmark: Olsen & Olsen; 1994. pp. 459–464.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

