Development of the piggyBac transposable system for Plasmodium berghei and its application for random mutagenesis in malaria parasites
- PMID: 21418605
- PMCID: PMC3073922
- DOI: 10.1186/1471-2164-12-155
Development of the piggyBac transposable system for Plasmodium berghei and its application for random mutagenesis in malaria parasites
Abstract
Background: The genome of a number of species of malaria parasites (Plasmodium spp.) has been sequenced in the hope of identifying new drug and vaccine targets. However, almost one-half of predicted Plasmodium genes are annotated as hypothetical and are difficult to analyse in bulk due to the inefficiency of current reverse genetic methodologies for Plasmodium. Recently, it has been shown that the transposase piggyBac integrates at random into the genome of the human malaria parasite P. falciparum offering the possibility to develop forward genetic screens to analyse Plasmodium gene function. This study reports the development and application of the piggyBac transposition system for the rodent malaria parasite P. berghei and the evaluation of its potential as a tool in forward genetic studies. P. berghei is the most frequently used malaria parasite model in gene function analysis since phenotype screens throughout the complete Plasmodium life cycle are possible both in vitro and in vivo.
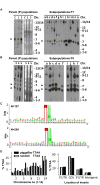
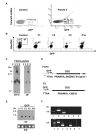
Results: We demonstrate that piggyBac based gene inactivation and promoter-trapping is both easier and more efficient in P. berghei than in the human malaria parasite, P. falciparum. Random piggyBac-mediated insertion into genes was achieved after parasites were transfected with the piggyBac donor plasmid either when transposase was expressed either from a helper plasmid or a stably integrated gene in the genome. Characterization of more than 120 insertion sites demonstrated that more than 70 most likely affect gene expression classifying their protein products as non-essential for asexual blood stage development. The non-essential nature of two of these genes was confirmed by targeted gene deletion one of which encodes P41, an ortholog of a human malaria vaccine candidate. Importantly for future development of whole genome phenotypic screens the remobilization of the piggyBac element in parasites that stably express transposase was demonstrated.
Conclusion: These data demonstrate that piggyBac behaved as an efficient and random transposon in P. berghei. Remobilization of piggyBac element shows that with further development the piggyBac system can be an effective tool to generate random genome-wide mutation parasite libraries, for use in large-scale phenotype screens in vitro and in vivo.
© 2011 Fonager et al; licensee BioMed Central Ltd.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

