Modeling antibiotic and cytotoxic effects of the dimeric isoquinoline IQ-143 on metabolism and its regulation in Staphylococcus aureus, Staphylococcus epidermidis and human cells
- PMID: 21418624
- PMCID: PMC3129674
- DOI: 10.1186/gb-2011-12-3-r24
Modeling antibiotic and cytotoxic effects of the dimeric isoquinoline IQ-143 on metabolism and its regulation in Staphylococcus aureus, Staphylococcus epidermidis and human cells
Abstract
Background: Xenobiotics represent an environmental stress and as such are a source for antibiotics, including the isoquinoline (IQ) compound IQ-143. Here, we demonstrate the utility of complementary analysis of both host and pathogen datasets in assessing bacterial adaptation to IQ-143, a synthetic analog of the novel type N,C-coupled naphthyl-isoquinoline alkaloid ancisheynine.
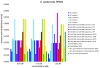
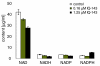
Results: Metabolite measurements, gene expression data and functional assays were combined with metabolic modeling to assess the effects of IQ-143 on Staphylococcus aureus, Staphylococcus epidermidis and human cell lines, as a potential paradigm for novel antibiotics. Genome annotation and PCR validation identified novel enzymes in the primary metabolism of staphylococci. Gene expression response analysis and metabolic modeling demonstrated the adaptation of enzymes to IQ-143, including those not affected by significant gene expression changes. At lower concentrations, IQ-143 was bacteriostatic, and at higher concentrations bactericidal, while the analysis suggested that the mode of action was a direct interference in nucleotide and energy metabolism. Experiments in human cell lines supported the conclusions from pathway modeling and found that IQ-143 had low cytotoxicity.
Conclusions: The data suggest that IQ-143 is a promising lead compound for antibiotic therapy against staphylococci. The combination of gene expression and metabolite analyses with in silico modeling of metabolite pathways allowed us to study metabolic adaptations in detail and can be used for the evaluation of metabolic effects of other xenobiotics.
Figures
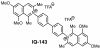



References
-
- Grundmann H, Aanensen DM, van den Wijngaard CC, Spratt BG, Harmsen D, Friedrich AW. the European Staphylococcal Reference Laboratory Working Group. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 2010;7:e1000215. doi: 10.1371/journal.pmed.1000215. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

