Genome-wide assessment of imprinted expression in human cells
- PMID: 21418647
- PMCID: PMC3129675
- DOI: 10.1186/gb-2011-12-3-r25
Genome-wide assessment of imprinted expression in human cells
Abstract
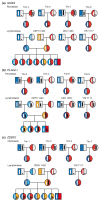
Background: Parent-of-origin-dependent expression of alleles, imprinting, has been suggested to impact a substantial proportion of mammalian genes. Its discovery requires allele-specific detection of expressed transcripts, but in some cases detected allelic expression bias has been interpreted as imprinting without demonstrating compatible transmission patterns and excluding heritable variation. Therefore, we utilized a genome-wide tool exploiting high density genotyping arrays in parallel measurements of genotypes in RNA and DNA to determine allelic expression across the transcriptome in lymphoblastoid cell lines (LCLs) and skin fibroblasts derived from families.
Results: We were able to validate 43% of imprinted genes with previous demonstration of compatible transmission patterns in LCLs and fibroblasts. In contrast, we only validated 8% of genes suggested to be imprinted in the literature, but without clear evidence of parent-of-origin-determined expression. We also detected five novel imprinted genes and delineated regions of imprinted expression surrounding annotated imprinted genes. More subtle parent-of-origin-dependent expression, or partial imprinting, could be verified in four genes. Despite higher prevalence of monoallelic expression, immortalized LCLs showed consistent imprinting in fewer loci than primary cells. Random monoallelic expression has previously been observed in LCLs and we show that random monoallelic expression in LCLs can be partly explained by aberrant methylation in the genome.
Conclusions: Our results indicate that widespread parent-of-origin-dependent expression observed recently in rodents is unlikely to be captured by assessment of human cells derived from adult tissues where genome-wide assessment of both primary and immortalized cells yields few new imprinted loci.
Figures
Comment in
-
The role of imprinted genes in humans.Genome Biol. 2011 Mar 21;12(3):106. doi: 10.1186/gb-2011-12-3-106. Genome Biol. 2011. PMID: 21418582 Free PMC article.
References
-
- Kong A, Steinthorsdottir V, Masson G, Thorleifsson G, Sulem P, Besenbacher S, Jonasdottir A, Sigurdsson A, Kristinsson KT, Jonasdottir A, Frigge ML, Gylfason A, Olason PI, Gudjonsson SA, Sverrisson S, Stacey SN, Sigurgeirsson B, Benediktsdottir KR, Sigurdsson H, Jonsson T, Benediktsson R, Olafsson JH, Johannsson OT, Hreidarsson AB, Sigurdsson G. DIAGRAM Consortium. Ferguson-Smith AC, Gudbjartsson DF, Thorsteinsdottir U, Stefansson K. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

