Role of the tail in the regulated state of myosin 2
- PMID: 21419133
- PMCID: PMC3776433
- DOI: 10.1016/j.jmb.2011.03.019
Role of the tail in the regulated state of myosin 2
Abstract
Myosin 2 from vertebrate smooth muscle or non-muscle sources is in equilibrium between compact, inactive monomers and thick filaments under physiological conditions. In the inactive monomer, the two heads pack compactly together, and the long tail is folded into three closely packed segments that are associated chiefly with one of the heads. The molecular basis of the folding of the tail remains unexplained. By using electron microscopy, we show that compact monomers of smooth muscle myosin 2 have the same structure in both the native state and following specific, intramolecular photo-cross-linking between Cys109 of the regulatory light chain (RLC) and segment 3 of the tail. Nonspecific cross-linking between lysine residues of the folded monomer by glutaraldehyde also does not perturb the compact conformation and stabilizes it against unfolding at high ionic strength. Sequence comparisons across phyla and myosin 2 isoforms suggest that the folding of the tail is stabilized by ionic interactions between the positively charged N-terminal sequence of the RLC and a negatively charged region near the start of tail segment 3 and that phosphorylation of the RLC could perturb these interactions. Our results support the view that interactions between the heads and the distal tail perform a critical role in regulating activity of myosin 2 molecules through stabilizing the compact monomer conformation.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
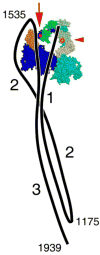
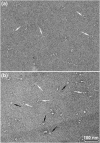








References
-
- Onishi H, Wakabayashi T, Kamata T, Watanabe S. Electron microscopic studies of myosin molecules from chicken gizzard muscle II: the effect of thiophosphorylation of the 20K-Dalton light chain on the ATP-induced change in the conformation of myosin monomers. J Biochem. 1983;94:1147–1154. - PubMed
-
- Trybus KM, Lowey S. Conformational states of smooth muscle myosin - Effects of light chain phosphorylation and ionic strength. J Biol Chem. 1984;259:8564–8571. - PubMed
-
- Craig R, Smith R, Kendrick-Jones J. Light-chain phosphorylation controls the conformation of vertebrate non-muscle and smooth muscle myosin molecules. Nature. 1983;302:436–439. - PubMed
-
- Ankrett RJ, Rowe AJ, Cross RA, Kendrick-Jones J, Bagshaw CR. A folded (10 S) conformer of myosin from a striated muscle and its implications for regulation of ATPase activity. J Mol Biol. 1991;217:323–335. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

