Microarray-based class discovery for molecular classification of breast cancer: analysis of interobserver agreement
- PMID: 21421860
- PMCID: PMC3079850
- DOI: 10.1093/jnci/djr071
Microarray-based class discovery for molecular classification of breast cancer: analysis of interobserver agreement
Abstract
Background: Breast cancers can be classified by hierarchical clustering using an "intrinsic" gene list into one of at least five molecular subtypes: basal-like, HER2, luminal A, luminal B, and normal breast-like. Five different intrinsic gene lists composed of varying numbers of genes have been used for molecular subtype identification and classification of breast cancers. The aim of this study was to determine the objectivity and interobserver reproducibility of the assignment of molecular subtype classes by hierarchical cluster analysis.
Methods: Three publicly available breast cancer datasets (n = 779) were subjected to two-way average-linkage hierarchical cluster analysis using five distinct intrinsic gene lists. We used free-marginal Kappa statistics to analyze interobserver agreement among five breast cancer researchers for the whole classification and for each molecular subtype separately according to each intrinsic gene list for each breast cancer dataset.
Results: None of the classification systems tested produced almost perfect agreement (Kappa ≥ 0.81) among observers. However, substantial interobserver agreement (70.8% to 76.1% of the samples and free-marginal Kappa scores from 0.635 to 0.701) was consistently observed in all datasets for four molecular subtypes (luminal, basal-like, HER2, and normal breast-like). When luminal cancers were subdivided (luminal A, B, and C), none of the classification systems produced substantial agreement (Kappa ≥ 0.61) in all the datasets analyzed. Analysis of each subtype separately revealed that only two (basal-like and HER2) could be reproducibly identified by independent observers (Kappa ≥ 0.81).
Conclusions: Assignment of molecular subtype classes of breast cancer based on the analysis of dendrograms obtained with hierarchical cluster analysis is subjective and shows modest interobserver reproducibility. For the development of a molecular taxonomy, objective definitions for each molecular subtype and standardized methods for their identification are required.
Figures
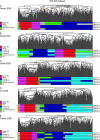
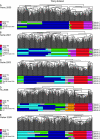
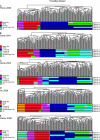
References
-
- Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. - PubMed
-
- Nielsen TO, West RB, Linn SC, et al. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 2002;359(9314):1301–1307. - PubMed
-
- Bittner M, Meltzer P, Chen Y, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406(6795):536–540. - PubMed
-
- Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

