Genetic basis of eye and pigment loss in the cave crustacean, Asellus aquaticus
- PMID: 21422298
- PMCID: PMC3078413
- DOI: 10.1073/pnas.1013850108
Genetic basis of eye and pigment loss in the cave crustacean, Asellus aquaticus
Abstract
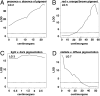
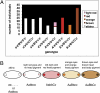
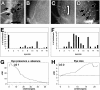
Understanding the process of evolution is one of the great challenges in biology. Cave animals are one group with immense potential to address the mechanisms of evolutionary change. Amazingly, similar morphological alterations, such as enhancement of sensory systems and the loss of eyes and pigmentation, have evolved multiple times in a diverse assemblage of cave animals. Our goal is to develop an invertebrate model to study cave evolution so that, in combination with a previously established vertebrate cave system, we can address genetic questions concerning evolutionary parallelism and convergence. We chose the isopod crustacean, Asellus aquaticus, and generated a genome-wide linkage map for this species. Our map, composed of 117 markers, of which the majority are associated with genes known to be involved in pigmentation, eye, and appendage development, was used to identify loci of large effect responsible for several pigmentation traits and eye loss. Our study provides support for the prediction that significant morphological change can be mediated through one or a few genes. Surprisingly, we found that within population variability in eye size occurs through multiple mechanisms; eye loss has a different genetic basis than reduced eye size. Similarly, again within a population, the phenotype of albinism can be achieved by two different genetic pathways--either by a recessive genotype at one locus or doubly recessive genotypes at two other loci. Our work shows the potential of Asellus for studying the extremes of parallel and convergent evolution-spanning comparisons within populations to comparisons between vertebrate and arthropod systems.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

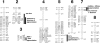
References
-
- Culver DC. Cave Life. Cambridge, MA: Harvard Univ Press; 1982.
-
- Wilkens H. Evolution and genetics of epigean and cave Astyanax (Characidae, Pisces) Evol Biol. 1988;23:271–367.
-
- Borowsky R, Wilkens H. Mapping a cave fish genome: polygenic systems and regressive evolution. J Hered. 2002;93:19–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

