Dominant prion mutants induce curing through pathways that promote chaperone-mediated disaggregation
- PMID: 21423195
- PMCID: PMC3082495
- DOI: 10.1038/nsmb.2031
Dominant prion mutants induce curing through pathways that promote chaperone-mediated disaggregation
Abstract
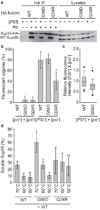
Protein misfolding underlies many neurodegenerative diseases, including the transmissible spongiform encephalopathies (prion diseases). Although cells typically recognize and process misfolded proteins, prion proteins evade protective measures by forming stable, self-replicating aggregates. However, coexpression of dominant-negative prion mutants can overcome aggregate accumulation and disease progression through currently unknown pathways. Here we determine the mechanisms by which two mutants of the Saccharomyces cerevisiae Sup35 protein cure the [PSI(+)] prion. We show that both mutants incorporate into wild-type aggregates and alter their physical properties in different ways, diminishing either their assembly rate or their thermodynamic stability. Whereas wild-type aggregates are recalcitrant to cellular intervention, mixed aggregates are disassembled by the molecular chaperone Hsp104. Thus, rather than simply blocking misfolding, dominant-negative prion mutants target multiple events in aggregate biogenesis to enhance their susceptibility to endogenous quality-control pathways.
Figures



Similar articles
-
The small heat shock protein Hsp31 cooperates with Hsp104 to modulate Sup35 prion aggregation.Prion. 2016 Nov;10(6):444-465. doi: 10.1080/19336896.2016.1234574. Prion. 2016. PMID: 27690738 Free PMC article.
-
DMSO-mediated curing of several yeast prion variants involves Hsp104 expression and protein solubilization, and is decreased in several autophagy related gene (atg) mutants.PLoS One. 2020 Mar 5;15(3):e0229796. doi: 10.1371/journal.pone.0229796. eCollection 2020. PLoS One. 2020. PMID: 32134970 Free PMC article.
-
A mathematical model of the dynamics of prion aggregates with chaperone-mediated fragmentation.J Math Biol. 2016 May;72(6):1555-78. doi: 10.1007/s00285-015-0921-0. Epub 2015 Aug 22. J Math Biol. 2016. PMID: 26297259 Free PMC article.
-
The story of stolen chaperones: how overexpression of Q/N proteins cures yeast prions.Prion. 2013 Jul-Aug;7(4):294-300. doi: 10.4161/pri.26021. Epub 2013 Aug 7. Prion. 2013. PMID: 23924684 Free PMC article. Review.
-
Three J-proteins impact Hsp104-mediated variant-specific prion elimination: a new critical role for a low-complexity domain.Curr Genet. 2020 Feb;66(1):51-58. doi: 10.1007/s00294-019-01006-5. Epub 2019 Jun 22. Curr Genet. 2020. PMID: 31230108 Free PMC article. Review.
Cited by
-
Effect of charged residues in the N-domain of Sup35 protein on prion [PSI+] stability and propagation.J Biol Chem. 2013 Oct 4;288(40):28503-13. doi: 10.1074/jbc.M113.471805. Epub 2013 Aug 21. J Biol Chem. 2013. PMID: 23965990 Free PMC article.
-
Loss of amino-terminal acetylation suppresses a prion phenotype by modulating global protein folding.Nat Commun. 2014 Jul 15;5:4383. doi: 10.1038/ncomms5383. Nat Commun. 2014. PMID: 25023910 Free PMC article.
-
Insights into prion biology: integrating a protein misfolding pathway with its cellular environment.Prion. 2011 Apr-Jun;5(2):76-83. doi: 10.4161/pri.5.2.16413. Epub 2011 Apr 1. Prion. 2011. PMID: 21654204 Free PMC article.
-
Prion propagation can occur in a prokaryote and requires the ClpB chaperone.Elife. 2014 Aug 13;3:e02949. doi: 10.7554/eLife.02949. Elife. 2014. PMID: 25122461 Free PMC article.
-
Nucleation seed size determines amyloid clearance and establishes a barrier to prion appearance in yeast.Nat Struct Mol Biol. 2020 Jun;27(6):540-549. doi: 10.1038/s41594-020-0416-6. Epub 2020 May 4. Nat Struct Mol Biol. 2020. PMID: 32367069 Free PMC article.
References
-
- Masel J, Jansen VA, Nowak MA. Quantifying the kinetic parameters of prion replication. Biophys Chem. 1999;77:139–152. - PubMed
-
- Collinge J, et al. Kuru in the 21st century--an acquired human prion disease with very long incubation periods. Lancet. 2006;367:2068–2074. - PubMed
-
- Gambetti P, Parchi P, Petersen RB, Chen SG, Lugaresi E. Fatal familial insomnia and familial Creutzfeldt-Jakob disease: clinical, pathological and molecular features. Brain Pathol. 1995;5:43–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

