The pro-longevity gene FoxO3 is a direct target of the p53 tumor suppressor
- PMID: 21423206
- PMCID: PMC3136551
- DOI: 10.1038/onc.2011.35
The pro-longevity gene FoxO3 is a direct target of the p53 tumor suppressor
Abstract
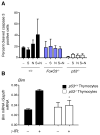
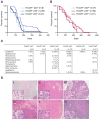
FoxO transcription factors have a conserved role in longevity, and act as tissue-specific tumor suppressors in mammals. Several nodes of interaction have been identified between FoxO transcription factors and p53, a major tumor suppressor in humans and mice. However, the extent and importance of the functional interaction between FoxO and p53 have not been fully explored. Here, we show that p53 regulates the expression of FoxO3, one of the four mammalian FoxO genes, in response to DNA damaging agents in both mouse embryonic fibroblasts and thymocytes. We find that p53 transactivates FoxO3 in cells by binding to a site in the second intron of the FoxO3 gene, a genomic region recently found to be associated with extreme longevity in humans. While FoxO3 is not necessary for p53-dependent cell cycle arrest, FoxO3 appears to modulate p53-dependent apoptosis. We also find that FoxO3 loss does not interact with p53 loss for tumor development in vivo, although the tumor spectrum of p53-deficient mice appears to be affected by FoxO3 loss. Our findings indicate that FoxO3 is a p53 target gene, and suggest that FoxO3 and p53 are part of a regulatory transcriptional network that may have an important role during aging and cancer.
Conflict of interest statement
We declare that there are no competing financial interests in relation to the work described.
Figures

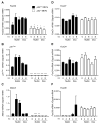
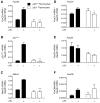

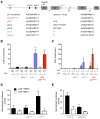
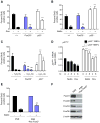
References
-
- Anselmi CV, Malovini A, Roncarati R, Novelli V, Villa F, Condorelli G, et al. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12:95–104. - PubMed
-
- Arum O, Johnson TE. Reduced expression of the Caenorhabditis elegans p53 ortholog cep-1 results in increased longevity. J Gerontol A Biol Sci Med Sci. 2007;62:951–9. - PubMed
-
- Bakker WJ, Harris IS, Mak TW. FOXO3a is activated in response to hypoxic stress and inhibits HIF1-induced apoptosis via regulation of CITED2. Mol Cell. 2007;28:941–53. - PubMed
-
- Bauer JH, Poon PC, Glatt-Deeley H, Abrams JM, Helfand SL. Neuronal expression of p53 dominant-negative proteins in adult Drosophila melanogaster extends life span. Curr Biol. 2005;15:2063–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

