Substoichiometrically different mitotypes coexist in mitochondrial genomes of Brassica napus L
- PMID: 21423700
- PMCID: PMC3053379
- DOI: 10.1371/journal.pone.0017662
Substoichiometrically different mitotypes coexist in mitochondrial genomes of Brassica napus L
Abstract
Cytoplasmic male sterility (CMS) has been identified in numerous plant species. Brassica napus CMS plants, such as Polima (pol), MI, and Shaan 2A, have been identified independently by different researchers with different materials in conventional breeding processes. How this kind of CMS emerges is unclear. Here, we report the mitochondrial genome sequence of the prevalent mitotype in the most widely used pol-CMS line, which has a length of 223,412 bp and encodes 34 proteins, 3 ribosomal RNAs, and 18 tRNAs, including two near identical copies of trnH. Of these 55 genes, 48 were found to be identical to their equivalents in the "nap" cytoplasm. The nap mitotype carries only one copy of trnH, and the sequences of five of the six remaining genes are highly similar to their equivalents in the pol mitotype. Forty-four open reading frames (ORFs) with unknown function were detected, including two unique to the pol mitotype (orf122 and orf132). At least five rearrangement events are required to account for the structural differences between the pol and nap sequences. The CMS-related orf224 neighboring region (∼5 kb) rearranged twice. PCR profiling based on mitotype-specific primer pairs showed that both mitotypes are present in B. napus cultivars. Quantitative PCR showed that the pol cytoplasm consists mainly of the pol mitotype, and the nap mitotype is the main genome of nap cytoplasm. Large variation in the copy number ratio of mitotypes was found, even among cultivars sharing the same cytoplasm. The coexistence of mitochondrial mitotypes and substoichiometric shifting can explain the emergence of CMS in B. napus.
Conflict of interest statement
Figures





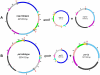
References
-
- McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–560. - PubMed
-
- Unseld M, Marienfeld JR, Brandt P, Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat Genet. 1997;15:57–61. - PubMed
-
- Satoh M, Kubo T, Nishizawa S, Estiati A, Itchoda N, et al. The cytoplasmic male-sterile type and normal type mitochondrial genomes of sugar beet share the same complement of genes of known function but differ in the content of expressed ORFs. Mol Genet Genomics. 2004;272:247–256. - PubMed
-
- Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, et al. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet Genomics. 2002;268:434–445. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

