To lyse or not to lyse: transient-mediated stochastic fate determination in cells infected by bacteriophages
- PMID: 21423715
- PMCID: PMC3053317
- DOI: 10.1371/journal.pcbi.1002006
To lyse or not to lyse: transient-mediated stochastic fate determination in cells infected by bacteriophages
Abstract
Cell fate determination is usually described as the result of the stochastic dynamics of gene regulatory networks (GRNs) reaching one of multiple steady-states each of which corresponds to a specific decision. However, the fate of a cell is determined in finite time suggesting the importance of transient dynamics in cellular decision making. Here we consider cellular decision making as resulting from first passage processes of regulatory proteins and examine the effect of transient dynamics within the initial lysis-lysogeny switch of phage λ. Importantly, the fate of an infected cell depends, in part, on the number of coinfecting phages. Using a quantitative model of the phage λ GRN, we find that changes in the likelihood of lysis and lysogeny can be driven by changes in phage co-infection number regardless of whether or not there exists steady-state bistability within the GRN. Furthermore, two GRNs which yield qualitatively distinct steady state behaviors as a function of phage infection number can show similar transient responses, sufficient for alternative cell fate determination. We compare our model results to a recent experimental study of cell fate determination in single cell assays of multiply infected bacteria. Whereas the experimental study proposed a "quasi-independent" hypothesis for cell fate determination consistent with an observed data collapse, we demonstrate that observed cell fate results are compatible with an alternative form of data collapse consistent with a partial gene dosage compensation mechanism. We show that including partial gene dosage compensation at the mRNA level in our stochastic model of fate determination leads to the same data collapse observed in the single cell study. Our findings elucidate the importance of transient gene regulatory dynamics in fate determination, and present a novel alternative hypothesis to explain single-cell level heterogeneity within the phage λ lysis-lysogeny decision switch.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

 and
and  while
while  and
and  is controlled by CII tetramers. Black arrows represent open reading frames of promoters when activated (
is controlled by CII tetramers. Black arrows represent open reading frames of promoters when activated ( ,
,  and
and  ) and antisense transcript aQ. (B) Interactions among gene products. Regular and blunt arrows represent positive and negative feedbacks, respectively. CI dimers are self-activators while repressing the other genes, and CRO dimers repress all the genes in the system. CII tetramers activate cI transcription, and suppress Q expression by transcribing antisense mRNAs.
) and antisense transcript aQ. (B) Interactions among gene products. Regular and blunt arrows represent positive and negative feedbacks, respectively. CI dimers are self-activators while repressing the other genes, and CRO dimers repress all the genes in the system. CII tetramers activate cI transcription, and suppress Q expression by transcribing antisense mRNAs.
 . Note that the system is bistable. (B) Phase diagram of CI-Q dynamics starting from no viral proteins when asymptotically divergent. Thresholds of CI and Q (both at 100 nM) represent the concentrations above which decisions are lysogenic and lytic, respectively. Trajectories cross the threshold only once. (C) Asymptotically divergent dynamics of Q concentration as a function of time. (D) Phase diagram of transiently divergent system with
. Note that the system is bistable. (B) Phase diagram of CI-Q dynamics starting from no viral proteins when asymptotically divergent. Thresholds of CI and Q (both at 100 nM) represent the concentrations above which decisions are lysogenic and lytic, respectively. Trajectories cross the threshold only once. (C) Asymptotically divergent dynamics of Q concentration as a function of time. (D) Phase diagram of transiently divergent system with  . Note that the system is not bistable. (E) Phase diagram of CI-Q dynamics of the transiently divergent phage
. Note that the system is not bistable. (E) Phase diagram of CI-Q dynamics of the transiently divergent phage  GRN. At
GRN. At  the deterministic trajectory crosses the threshold three times, and decisions change from lysis to lysogeny as a function of time. (F) Transiently divergent Q dynamics.
the deterministic trajectory crosses the threshold three times, and decisions change from lysis to lysogeny as a function of time. (F) Transiently divergent Q dynamics.

 and
and  represent the number of coinfecting phages and the host cell volume, respectively, so
represent the number of coinfecting phages and the host cell volume, respectively, so  is the phage genome concentration. Each point is the result from 5,000 simulations.
is the phage genome concentration. Each point is the result from 5,000 simulations.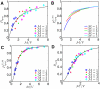
 . Each phage within a host is completely independent from other phages, and decision of lysogeny becomes a function of host volume. Note that rescaled curves do not collapse into a single curve. (C) Rescaled probability of
. Each phage within a host is completely independent from other phages, and decision of lysogeny becomes a function of host volume. Note that rescaled curves do not collapse into a single curve. (C) Rescaled probability of  proposed by Zeng et. al.
representing the probability of lysogeny for each individual infecting phage. Each phage independently “chooses” lysis or lysogeny. However, since the fraction of lysogeny for a single phage is a function of
proposed by Zeng et. al.
representing the probability of lysogeny for each individual infecting phage. Each phage independently “chooses” lysis or lysogeny. However, since the fraction of lysogeny for a single phage is a function of  , phages sense the presence of other phages. Note that data from different
, phages sense the presence of other phages. Note that data from different  -s collapse into a single curve. (D) Probability of lysogeny plotted against rescaled
-s collapse into a single curve. (D) Probability of lysogeny plotted against rescaled  when
when  , corresponding to a mechanism in which gene expression from multiple copies is partially compensated. Due to partial dosage compensation, the transcription rate is not linearly proportional to
, corresponding to a mechanism in which gene expression from multiple copies is partially compensated. Due to partial dosage compensation, the transcription rate is not linearly proportional to  , and the effective copy number is given as
, and the effective copy number is given as  where
where  . Note that the data from different
. Note that the data from different  -s collapse into a single curve. Black lines represent nonlinear curve fits into Hill functions.
-s collapse into a single curve. Black lines represent nonlinear curve fits into Hill functions.
 dependence as seen in the experimental data (see Fig. 5). (B) Simulation results on the fraction of lysogeny from Fig. 6 (A) plotted with rescaled
dependence as seen in the experimental data (see Fig. 5). (B) Simulation results on the fraction of lysogeny from Fig. 6 (A) plotted with rescaled  when
when  . The outcome of stochastic simulations with partial dosage compensation is consistent with experimental data (see Fig. 5 (A,D)). In this case, the GRN is asymptotically driven with CI and Q threshold at 100 nM and 120 nM, respectively, all other parameters are set according to Table 1 - transiently divergent. Each point is the result from 3,000 simulations.
. The outcome of stochastic simulations with partial dosage compensation is consistent with experimental data (see Fig. 5 (A,D)). In this case, the GRN is asymptotically driven with CI and Q threshold at 100 nM and 120 nM, respectively, all other parameters are set according to Table 1 - transiently divergent. Each point is the result from 3,000 simulations.Similar articles
-
From Bench to Keyboard and Back Again: A Brief History of Lambda Phage Modeling.Annu Rev Biophys. 2021 May 6;50:117-134. doi: 10.1146/annurev-biophys-082020-063558. Annu Rev Biophys. 2021. PMID: 33957052 Free PMC article. Review.
-
Stochastic cellular fate decision making by multiple infecting lambda phage.PLoS One. 2014 Aug 8;9(8):e103636. doi: 10.1371/journal.pone.0103636. eCollection 2014. PLoS One. 2014. PMID: 25105971 Free PMC article.
-
Interactions between Viral Regulatory Proteins Ensure an MOI-Independent Probability of Lysogeny during Infection by Bacteriophage P1.mBio. 2021 Oct 26;12(5):e0101321. doi: 10.1128/mBio.01013-21. Epub 2021 Sep 14. mBio. 2021. PMID: 34517752 Free PMC article.
-
Minimal gene regulatory circuits for a lysis-lysogeny choice in the presence of noise.PLoS One. 2010 Dec 20;5(12):e15037. doi: 10.1371/journal.pone.0015037. PLoS One. 2010. PMID: 21188148 Free PMC article.
-
High-resolution studies of lysis-lysogeny decision-making in bacteriophage lambda.J Biol Chem. 2019 Mar 8;294(10):3343-3349. doi: 10.1074/jbc.TM118.003209. Epub 2018 Sep 21. J Biol Chem. 2019. PMID: 30242122 Free PMC article. Review.
Cited by
-
Identification of control targets in Boolean molecular network models via computational algebra.BMC Syst Biol. 2016 Sep 23;10(1):94. doi: 10.1186/s12918-016-0332-x. BMC Syst Biol. 2016. PMID: 27662842 Free PMC article.
-
Phage-inducible chromosomal islands promote genetic variability by blocking phage reproduction and protecting transductants from phage lysis.PLoS Genet. 2022 Mar 28;18(3):e1010146. doi: 10.1371/journal.pgen.1010146. eCollection 2022 Mar. PLoS Genet. 2022. PMID: 35344558 Free PMC article.
-
Life-style and genome structure of marine Pseudoalteromonas siphovirus B8b isolated from the northwestern Mediterranean Sea.PLoS One. 2015 Jan 14;10(1):e0114829. doi: 10.1371/journal.pone.0114829. eCollection 2015. PLoS One. 2015. PMID: 25587991 Free PMC article.
-
From Bench to Keyboard and Back Again: A Brief History of Lambda Phage Modeling.Annu Rev Biophys. 2021 May 6;50:117-134. doi: 10.1146/annurev-biophys-082020-063558. Annu Rev Biophys. 2021. PMID: 33957052 Free PMC article. Review.
-
Stochastic cellular fate decision making by multiple infecting lambda phage.PLoS One. 2014 Aug 8;9(8):e103636. doi: 10.1371/journal.pone.0103636. eCollection 2014. PLoS One. 2014. PMID: 25105971 Free PMC article.
References
-
- Solomon JM, Grossman AD. Who's competent and when: regulation of natural genetic competence in bacteria. Trends Genet. 1996;12:150–155. - PubMed
-
- Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Gen. 1996;30:297–341. - PubMed
-
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. - PubMed
-
- Ptashne M. Phage lambda revisted. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 3rd edition; 2004. A genetic switch.
-
- Morrison SJ, Shar NM. Regulatory mechanisms in stem cell biology. Cell. 1997;88:287–298. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

