The role of incoherent microRNA-mediated feedforward loops in noise buffering
- PMID: 21423718
- PMCID: PMC3053320
- DOI: 10.1371/journal.pcbi.1001101
The role of incoherent microRNA-mediated feedforward loops in noise buffering
Abstract
MicroRNAs are endogenous non-coding RNAs which negatively regulate the expression of protein-coding genes in plants and animals. They are known to play an important role in several biological processes and, together with transcription factors, form a complex and highly interconnected regulatory network. Looking at the structure of this network, it is possible to recognize a few overrepresented motifs which are expected to perform important elementary regulatory functions. Among them, a special role is played by the microRNA-mediated feedforward loop in which a master transcription factor regulates a microRNA and, together with it, a set of target genes. In this paper we show analytically and through simulations that the incoherent version of this motif can couple the fine-tuning of a target protein level with an efficient noise control, thus conferring precision and stability to the overall gene expression program, especially in the presence of fluctuations in upstream regulators. Among the other results, a nontrivial prediction of our model is that the optimal attenuation of fluctuations coincides with a modest repression of the target expression. This feature is coherent with the expected fine-tuning function and in agreement with experimental observations of the actual impact of a wide class of microRNAs on the protein output of their targets. Finally, we describe the impact on noise-buffering efficiency of the cross-talk between microRNA targets that can naturally arise if the microRNA-mediated circuit is not considered as isolated, but embedded in a larger network of regulations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

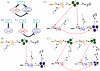
 ) are transcribed and eventually degraded (broken lines). RNAs can be translated into proteins (
) are transcribed and eventually degraded (broken lines). RNAs can be translated into proteins ( is the TF while
is the TF while  is the target protein) symbolized by circles, and proteins can be degraded (broken circles). Rates of each process (transcription, translation or degradation) are depicted along the corresponding black arrows. Regulations are represented in red, with arrows in the case of activation by TFs while rounded end lines in the case of miRNA repression. TF regulations act on rates of transcription that become functions of the amount of regulators. MiRNA regulation makes the rate of translation of the target a function of miRNA concentration.
is the target protein) symbolized by circles, and proteins can be degraded (broken circles). Rates of each process (transcription, translation or degradation) are depicted along the corresponding black arrows. Regulations are represented in red, with arrows in the case of activation by TFs while rounded end lines in the case of miRNA repression. TF regulations act on rates of transcription that become functions of the amount of regulators. MiRNA regulation makes the rate of translation of the target a function of miRNA concentration.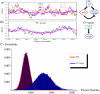
 and of translation
and of translation  . Proteins degrade with a rate
. Proteins degrade with a rate  , while mRNAs and miRNAs with
, while mRNAs and miRNAs with  . The parameters in the Hill functions of regulation (equations 1,2) are the following: the maximum rate of transcription for mRNAs is
. The parameters in the Hill functions of regulation (equations 1,2) are the following: the maximum rate of transcription for mRNAs is  , while for miRNAs is
, while for miRNAs is  ; the maximum rate of translation of the target is
; the maximum rate of translation of the target is  ; dissociation constants are
; dissociation constants are  ; Hill coefficients are all
; Hill coefficients are all  , as typical biological values range from 1 (hyperbolic control) to 30 (sharp switching). (B) Time evolution in a simulation for the molecular players in the linear TF-gene cascade (scheme on the right or more detailed in Figure 2B′). Compared to the FFL case, the
, as typical biological values range from 1 (hyperbolic control) to 30 (sharp switching). (B) Time evolution in a simulation for the molecular players in the linear TF-gene cascade (scheme on the right or more detailed in Figure 2B′). Compared to the FFL case, the  evolution is more sensitive to TF fluctuations. (C) The probability distribution of protein number for the two circuits. Histograms are the result of Gillespie simulations while continuous lines are empirical distributions (gaussian for the FFL and gamma for the TF-gene) with mean and variance predicted by the analytical model.
evolution is more sensitive to TF fluctuations. (C) The probability distribution of protein number for the two circuits. Histograms are the result of Gillespie simulations while continuous lines are empirical distributions (gaussian for the FFL and gamma for the TF-gene) with mean and variance predicted by the analytical model.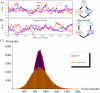
 and
and  trajectories that is present in the FFL (A) is completely lost in the open circuit. As a consequence while the mean value of
trajectories that is present in the FFL (A) is completely lost in the open circuit. As a consequence while the mean value of  is approximately the same, its fluctuations are appreciably greater in the open circuit case. (C) The probability distribution of protein number for the two circuits. Histograms are the result of Gillespie simulations while continuous lines are empirical distributions (gaussian for the FFL and gamma for the open circuit) with mean and variance predicted by the analytical model.
is approximately the same, its fluctuations are appreciably greater in the open circuit case. (C) The probability distribution of protein number for the two circuits. Histograms are the result of Gillespie simulations while continuous lines are empirical distributions (gaussian for the FFL and gamma for the open circuit) with mean and variance predicted by the analytical model.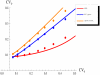
 , while we vary its relative fluctuations
, while we vary its relative fluctuations  , tuning the relative contribution of transcription (rate
, tuning the relative contribution of transcription (rate  ) and translation (rate
) and translation (rate  ). All the other parameters have the values reported in caption of Figure 3. The incoherent FFL makes the target less sensitive to fluctuations in the upstream TF. The extent of the noise reduction, with respect to the other circuits, depends on the magnitude of the TF noise, pointing out that the FFL topology is particularly effective in filtering out extrinsic fluctuations. Dots are the result of Gillespie simulations with the full nonlinear dynamics while continuous lines are analytical predictions.
). All the other parameters have the values reported in caption of Figure 3. The incoherent FFL makes the target less sensitive to fluctuations in the upstream TF. The extent of the noise reduction, with respect to the other circuits, depends on the magnitude of the TF noise, pointing out that the FFL topology is particularly effective in filtering out extrinsic fluctuations. Dots are the result of Gillespie simulations with the full nonlinear dynamics while continuous lines are analytical predictions.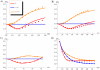
 as a function of the repression strength
as a function of the repression strength  . The Figure shows the presence of an optimal repression strength for which the introduction of a miRNA regulation in a FFL minimizes noise. (B)
. The Figure shows the presence of an optimal repression strength for which the introduction of a miRNA regulation in a FFL minimizes noise. (B)  as a function of the mean number of miRNAs
as a function of the mean number of miRNAs  . In this case
. In this case  is changed through the maximum rate of transcription
is changed through the maximum rate of transcription  (see equation 1). (C)
(see equation 1). (C)  as a function of
as a function of  , varying the dissociation constant
, varying the dissociation constant  . In both cases (B and C) is evident a U-shaped profile, implying an optimal noise buffering for intermediate miRNA concentrations. (D)
. In both cases (B and C) is evident a U-shaped profile, implying an optimal noise buffering for intermediate miRNA concentrations. (D)  as a function of the mean number of TFs
as a function of the mean number of TFs  . The number of TFs depends on the rate of their transcription
. The number of TFs depends on the rate of their transcription  and of their translation
and of their translation  . The Figure is obtained manipulating
. The Figure is obtained manipulating  , but the alternative choice of
, but the alternative choice of  leads to equivalent results (see Text S1). For intermediate concentration of the TF, the noise control by the FFL outperforms the one of the other circuits. In each plot, dots are the result of Gillespie simulations while continuous lines are analytical predictions. The values of parameters kept constant are the same of Figure 3, however the results are quite robust with respect to their choice (see Text S1 for details).
leads to equivalent results (see Text S1). For intermediate concentration of the TF, the noise control by the FFL outperforms the one of the other circuits. In each plot, dots are the result of Gillespie simulations while continuous lines are analytical predictions. The values of parameters kept constant are the same of Figure 3, however the results are quite robust with respect to their choice (see Text S1 for details).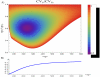
 , achieved with the FFL, is evaluated with respect to noise deriving from constitutive expression
, achieved with the FFL, is evaluated with respect to noise deriving from constitutive expression  (i.e. in absence of miRNA regulation) for different mean levels of the TF
(i.e. in absence of miRNA regulation) for different mean levels of the TF  and different degrees of reduction of the target protein level
and different degrees of reduction of the target protein level  (where
(where  is the mean constitutive expression). The TF level is changed through its rate of translation
is the mean constitutive expression). The TF level is changed through its rate of translation  (equivalent results can be obtained changing the rate of transcription
(equivalent results can be obtained changing the rate of transcription  ), while the target level is tuned varying the repression strength. All the other parameters have the values reported in caption of Figure 3 except
), while the target level is tuned varying the repression strength. All the other parameters have the values reported in caption of Figure 3 except  (lower than in Figure 3 to explore a more noisy situation). The region where miRNA repression leads to larger fluctuations with respect to constitutive ones is shown in white. When a noise reduction is gained the value of
(lower than in Figure 3 to explore a more noisy situation). The region where miRNA repression leads to larger fluctuations with respect to constitutive ones is shown in white. When a noise reduction is gained the value of  is reported with the color code explained in the legend. The best noise control is achieved with a modest suppression of target expression, around the 60% of its constitutive value. (B) The rate of transcription of the target mRNA as a function of the mean number of TFs. The noise reduction shown in the above plot can be obtained outside the saturation regime (where the slope of the activation curve tends to zero).
is reported with the color code explained in the legend. The best noise control is achieved with a modest suppression of target expression, around the 60% of its constitutive value. (B) The rate of transcription of the target mRNA as a function of the mean number of TFs. The noise reduction shown in the above plot can be obtained outside the saturation regime (where the slope of the activation curve tends to zero).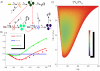
 as a function of the repression strength
as a function of the repression strength  for a miRNA-mediated FFL and for its transcriptional counterpart. Thanks to the constraints imposed on parameters we can directly compare their noise-buffering efficiency with respect to a gene only activated by a TF. Both circuitries lead to a
for a miRNA-mediated FFL and for its transcriptional counterpart. Thanks to the constraints imposed on parameters we can directly compare their noise-buffering efficiency with respect to a gene only activated by a TF. Both circuitries lead to a  curve with a minimum for an intermediate repression strength, but the miRNA-mediated circuit appears more efficient in filtering out fluctuations. The values of parameters kept constant are the same of Figure 3. Dots are the result of Gillespie simulations with the full nonlinear dynamics while continuous lines are analytical predictions. Also in this case, analytical solutions fit nicely with simulation results. (C) The noise reduction
curve with a minimum for an intermediate repression strength, but the miRNA-mediated circuit appears more efficient in filtering out fluctuations. The values of parameters kept constant are the same of Figure 3. Dots are the result of Gillespie simulations with the full nonlinear dynamics while continuous lines are analytical predictions. Also in this case, analytical solutions fit nicely with simulation results. (C) The noise reduction  , achieved with a purely transcriptional incoherent FFL, evaluated for different mean levels of the TF
, achieved with a purely transcriptional incoherent FFL, evaluated for different mean levels of the TF  and different degrees of reduction of the target protein level
and different degrees of reduction of the target protein level  . The parameter values and the color code are the same of Figure 7 so as to allow a direct comparison.
. The parameter values and the color code are the same of Figure 7 so as to allow a direct comparison.
 is depicted as a function of the ratio of effective transcription rates of the secondary target (
is depicted as a function of the ratio of effective transcription rates of the secondary target ( ) and of the FFL joint target (
) and of the FFL joint target ( ), for different values of
), for different values of  . Since the rate of transcription of the joint target is a function of the TF concentration, we consider for this analysis the effective mean rate
. Since the rate of transcription of the joint target is a function of the TF concentration, we consider for this analysis the effective mean rate  as a reference (where
as a reference (where  is constant as we are not tuning the TF concentration). The transcription of the second target is modeled as an independent birth-death process with birth rate
is constant as we are not tuning the TF concentration). The transcription of the second target is modeled as an independent birth-death process with birth rate  . In this plot the coupling constants between targets and miRNAs are assumed equal (
. In this plot the coupling constants between targets and miRNAs are assumed equal ( ) and for each
) and for each  value the coupling constant
value the coupling constant  is chosen so as to start with the same amount of target proteins (
is chosen so as to start with the same amount of target proteins ( ) in absence of secondary targets (the complete set of parameters values is presented in Text S1). In the limit of infinite out-of-circuit target expression, the joint target protein level approaches its constitutive value if
) in absence of secondary targets (the complete set of parameters values is presented in Text S1). In the limit of infinite out-of-circuit target expression, the joint target protein level approaches its constitutive value if  , while remains constant in the ideal case of perfectly catalytic miRNA repression (red curve). Continuous lines are analytical solutions of the deterministic model (Equations 9), while dots are the result of stochastic simulations. (C) With the parameter setting of Figure 9B, the noise reduction
, while remains constant in the ideal case of perfectly catalytic miRNA repression (red curve). Continuous lines are analytical solutions of the deterministic model (Equations 9), while dots are the result of stochastic simulations. (C) With the parameter setting of Figure 9B, the noise reduction  is evaluated in the same
is evaluated in the same  range. Dots are the result of Gillespie simulations while continuous lines come from trivial interpolations. (D) The noise reduction is evaluated as a function of the out-of-circuit mRNA fluctuations
range. Dots are the result of Gillespie simulations while continuous lines come from trivial interpolations. (D) The noise reduction is evaluated as a function of the out-of-circuit mRNA fluctuations  , relative to the joint target fluctuations
, relative to the joint target fluctuations  . The fluctuations of the second target are modulated considering its rate of transcription as a function of an independent TF and changing the TF noise with the same strategy used for Figure 5 (see Text S1 for more details). The concentrations of the TFs activating the two targets are constrained to be equal so as to study the situation of two independent targets with the same effective transcription rate. Dots are the result of Gillespie simulations, simply interpolated with continuous lines.
. The fluctuations of the second target are modulated considering its rate of transcription as a function of an independent TF and changing the TF noise with the same strategy used for Figure 5 (see Text S1 for more details). The concentrations of the TFs activating the two targets are constrained to be equal so as to study the situation of two independent targets with the same effective transcription rate. Dots are the result of Gillespie simulations, simply interpolated with continuous lines.References
-
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Bushati N, Cohen SN. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. - PubMed
-
- Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

