Dynamic changes in the microRNA expression profile reveal multiple regulatory mechanisms in the spinal nerve ligation model of neuropathic pain
- PMID: 21423802
- PMCID: PMC3056716
- DOI: 10.1371/journal.pone.0017670
Dynamic changes in the microRNA expression profile reveal multiple regulatory mechanisms in the spinal nerve ligation model of neuropathic pain
Abstract
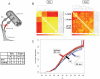
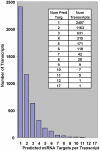
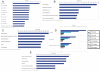
Neuropathic pain resulting from nerve lesions or dysfunction represents one of the most challenging neurological diseases to treat. A better understanding of the molecular mechanisms responsible for causing these maladaptive responses can help develop novel therapeutic strategies and biomarkers for neuropathic pain. We performed a miRNA expression profiling study of dorsal root ganglion (DRG) tissue from rats four weeks post spinal nerve ligation (SNL), a model of neuropathic pain. TaqMan low density arrays identified 63 miRNAs whose level of expression was significantly altered following SNL surgery. Of these, 59 were downregulated and the ipsilateral L4 DRG, not the injured L5 DRG, showed the most significant downregulation suggesting that miRNA changes in the uninjured afferents may underlie the development and maintenance of neuropathic pain. TargetScan was used to predict mRNA targets for these miRNAs and it was found that the transcripts with multiple predicted target sites belong to neurologically important pathways. By employing different bioinformatic approaches we identified neurite remodeling as a significantly regulated biological pathway, and some of these predictions were confirmed by siRNA knockdown for genes that regulate neurite growth in differentiated Neuro2A cells. In vitro validation for predicted target sites in the 3'-UTR of voltage-gated sodium channel Scn11a, alpha 2/delta1 subunit of voltage-dependent Ca-channel, and purinergic receptor P2rx ligand-gated ion channel 4 using luciferase reporter assays showed that identified miRNAs modulated gene expression significantly. Our results suggest the potential for miRNAs to play a direct role in neuropathic pain.
Conflict of interest statement
Figures
Similar articles
-
Voltage-gated sodium channel 1.7 expression decreases in dorsal root ganglia in a spinal nerve ligation neuropathic pain model.Kaohsiung J Med Sci. 2019 Aug;35(8):493-500. doi: 10.1002/kjm2.12088. Epub 2019 May 14. Kaohsiung J Med Sci. 2019. PMID: 31087766 Free PMC article.
-
Impaired neuropathic pain and preserved acute pain in rats overexpressing voltage-gated potassium channel subunit Kv1.2 in primary afferent neurons.Mol Pain. 2014 Jan 29;10:8. doi: 10.1186/1744-8069-10-8. Mol Pain. 2014. PMID: 24472174 Free PMC article.
-
Circulating microRNA expression profile: a novel potential predictor for chronic nervous lesions.Acta Biochim Biophys Sin (Shanghai). 2014 Nov;46(11):942-9. doi: 10.1093/abbs/gmu090. Epub 2014 Sep 30. Acta Biochim Biophys Sin (Shanghai). 2014. PMID: 25274330
-
Protein Kinases as Mediators for miRNA Modulation of Neuropathic Pain.Cells. 2025 Apr 11;14(8):577. doi: 10.3390/cells14080577. Cells. 2025. PMID: 40277902 Free PMC article. Review.
-
Ion channel long non-coding RNAs in neuropathic pain.Pflugers Arch. 2022 Apr;474(4):457-468. doi: 10.1007/s00424-022-02675-x. Epub 2022 Mar 2. Pflugers Arch. 2022. PMID: 35235008 Review.
Cited by
-
microRNAs in nociceptive circuits as predictors of future clinical applications.Front Mol Neurosci. 2013 Oct 17;6:33. doi: 10.3389/fnmol.2013.00033. Front Mol Neurosci. 2013. PMID: 24151455 Free PMC article. Review.
-
Increased miR-132-3p expression is associated with chronic neuropathic pain.Exp Neurol. 2016 Sep;283(Pt A):276-86. doi: 10.1016/j.expneurol.2016.06.025. Epub 2016 Jun 24. Exp Neurol. 2016. PMID: 27349406 Free PMC article.
-
Intrathecal miR-96 inhibits Nav1.3 expression and alleviates neuropathic pain in rat following chronic construction injury.Neurochem Res. 2014 Jan;39(1):76-83. doi: 10.1007/s11064-013-1192-z. Neurochem Res. 2014. PMID: 24234845
-
Dicer-microRNA pathway is critical for peripheral nerve regeneration and functional recovery in vivo and regenerative axonogenesis in vitro.Exp Neurol. 2012 Jan;233(1):555-65. doi: 10.1016/j.expneurol.2011.11.041. Epub 2011 Dec 8. Exp Neurol. 2012. PMID: 22178326 Free PMC article.
-
Dynamic and differential regulation in the microRNA expression in the developing and mature cataractous rat lens.J Cell Mol Med. 2013 Sep;17(9):1146-59. doi: 10.1111/jcmm.12094. Epub 2013 Jul 11. J Cell Mol Med. 2013. PMID: 23844765 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

