Genetic characteristics, coreceptor usage potential and evolution of Nigerian HIV-1 subtype G and CRF02_AG isolates
- PMID: 21423811
- PMCID: PMC3056731
- DOI: 10.1371/journal.pone.0017865
Genetic characteristics, coreceptor usage potential and evolution of Nigerian HIV-1 subtype G and CRF02_AG isolates
Abstract
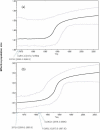
HIV-1 CRF02_AG and subtype G (HIV-1G) account for most HIV infections in Nigeria, but their evolutionary trends have not been well documented. To better elucidate the dynamics of the epidemic in Nigeria we characterised the gag and env genes of North-Central Nigerian HIV-1 isolates from pregnant women. Of 28 samples sequenced in both genes, the predominant clades were CRF02_AG (39%) and HIV-1G (32%). Higher predicted proportion of CXCR4-tropic (X4) HIV-1G isolates was noted compared to CRF02_AG (p = 0.007, Fisher's exact test). Phylogenetic and Bayesian analysis conducted on our sequences and all the dated available Nigerian sequences on the Los Alamos data base showed that CRF02_AG and HIV-1G entered into Nigeria through multiple entries, with presence of HIV-1G dating back to early 1980s. This study underlines the genetic complexity of the HIV-1 epidemic in Nigeria, possible subtype-specific differences in co-receptor usage, and the evolutionary trends of the predominant HIV-1 strains in Nigeria, which may have implications for the design of biomedical interventions and better understanding of the epidemic.
Conflict of interest statement
Figures


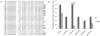

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

