Fast extraction of neuron morphologies from large-scale SBFSEM image stacks
- PMID: 21424815
- PMCID: PMC3232351
- DOI: 10.1007/s10827-011-0316-1
Fast extraction of neuron morphologies from large-scale SBFSEM image stacks
Abstract
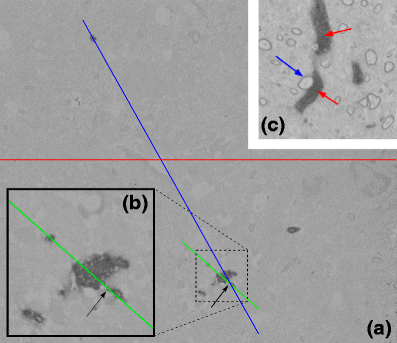
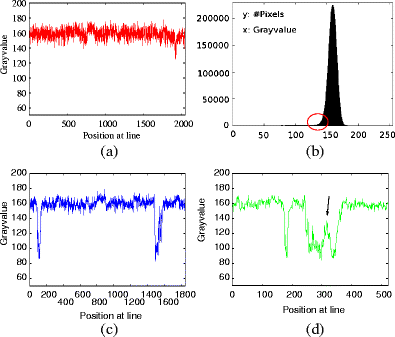
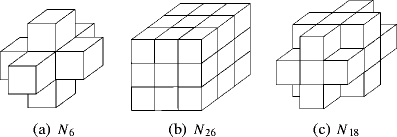
Neuron morphology is frequently used to classify cell-types in the mammalian cortex. Apart from the shape of the soma and the axonal projections, morphological classification is largely defined by the dendrites of a neuron and their subcellular compartments, referred to as dendritic spines. The dimensions of a neuron's dendritic compartment, including its spines, is also a major determinant of the passive and active electrical excitability of dendrites. Furthermore, the dimensions of dendritic branches and spines change during postnatal development and, possibly, following some types of neuronal activity patterns, changes depending on the activity of a neuron. Due to their small size, accurate quantitation of spine number and structure is difficult to achieve (Larkman, J Comp Neurol 306:332, 1991). Here we follow an analysis approach using high-resolution EM techniques. Serial block-face scanning electron microscopy (SBFSEM) enables automated imaging of large specimen volumes at high resolution. The large data sets generated by this technique make manual reconstruction of neuronal structure laborious. Here we present NeuroStruct, a reconstruction environment developed for fast and automated analysis of large SBFSEM data sets containing individual stained neurons using optimized algorithms for CPU and GPU hardware. NeuroStruct is based on 3D operators and integrates image information from image stacks of individual neurons filled with biocytin and stained with osmium tetroxide. The focus of the presented work is the reconstruction of dendritic branches with detailed representation of spines. NeuroStruct delivers both a 3D surface model of the reconstructed structures and a 1D geometrical model corresponding to the skeleton of the reconstructed structures. Both representations are a prerequisite for analysis of morphological characteristics and simulation signalling within a neuron that capture the influence of spines.
Figures





References
-
- Adams R, Bischof L. Seeded region growing. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1994;16(6):641–647. doi: 10.1109/34.295913. - DOI
-
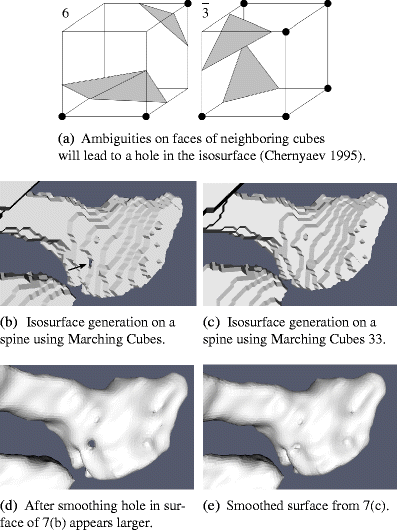
- Chernyaev, E. V. (1995). Marching cubes 33: Construction of topologically correct isosurfaces. Tech. rep., CERN CN 95-17.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

