Characterization and comparison of disulfide linkages and scrambling patterns in therapeutic monoclonal antibodies: using LC-MS with electron transfer dissociation
- PMID: 21428412
- PMCID: PMC3082428
- DOI: 10.1021/ac200128d
Characterization and comparison of disulfide linkages and scrambling patterns in therapeutic monoclonal antibodies: using LC-MS with electron transfer dissociation
Abstract
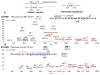
The disulfides in three monoclonal antibodies (mAb), the anti-HER2, anti-CD11a, and GLP-1 with IgG4-Fc fusion protein, were completely mapped by LC-MS with the combination of electron-transfer dissociation (ETD) and collision induced dissociation (CID) fragmentation. In addition to mapping the 4 inter- and 12 intrachain disulfides (total 16), the identification of scrambled disulfides in degraded samples (heat-stress) was achieved. The scrambling was likely attributed to an initial breakage between the light (Cys 214) and heavy (Cys 223) chains in anti-HER2, with the same observation found in a similar therapeutic mAb, anti-CD11a. On the other hand, the fusion antibody, with no light chain but containing only two heavy chains, generated much less scrambling under the same heat-stressed conditions. The preferred sites of scrambling were identified, such as the intrachain disulfide for CxxC in the heavy chain, and the C194 of the heavy chain pairing with the terminal Cys residue (C214) in the light chain. The interchain disulfides between the light and heavy chains were weaker than the interchain disulfides between the two heavy chains. The relative high abundance ions observed in ETD provided strong evidence for the linked peptide information, which was particularly useful for the identification of the scrambled disulfides. The use of sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) helped the separation of these misfolded proteins for the determination of scrambled disulfide linkages. This methodology is useful for comparison of disulfide stability generated from different structural designs and providing a new way to determine the scrambling patterns, which could be applied for those seeking to determine unknown disulfide linkages.
© 2011 American Chemical Society
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

