Histone methylases MLL1 and MLL3 coordinate with estrogen receptors in estrogen-mediated HOXB9 expression
- PMID: 21428455
- PMCID: PMC3082634
- DOI: 10.1021/bi102037t
Histone methylases MLL1 and MLL3 coordinate with estrogen receptors in estrogen-mediated HOXB9 expression
Abstract
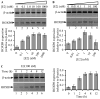
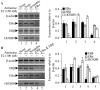
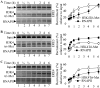
Homeobox gene HOXB9 is a critical player in development of mammary gland and sternum and in regulation of renin which is closely linked with blood pressure control. Our studies demonstrated that HOXB9 gene is transcriptionally regulated by estrogen (E2). HOXB9 promoter contains several estrogen-response elements (ERE). Reporter assay based experiments demonstrated that HOXB9 promoter EREs are estrogen responsive. Estrogen receptors ERα and ERβ are essential for E2-mediated transcriptional activation of HOXB9. Chromatin immunoprecipitation assay demonstrated that ERs bind to HOXB9 EREs as a function of E2. Similarly, histone methylases MLL1 and MLL3 also bind to HOXB9 EREs and play a critical role in E2-mediated transcriptional activation of HOXB9. Overall, our studies demonstrated that HOXB9 is an E2-responsive gene and ERs coordinate with MLL1 and MLL3 in E2-mediated transcriptional regulation of HOXB9.
Figures





References
-
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. - PubMed
-
- Ansari KI, Mishra BP, Mandal SS. Human CpG binding protein interacts with MLL1, MLL2 and hSet1 and regulates Hox gene expression. Biochim Biophys Acta. 2008;1779:66–73. - PubMed
-
- Ansari KI, Mishra BP, Mandal SS. MLL histone methylases in gene expression, hormone signaling and cell cycle. Front Biosci. 2009;14:3483–3495. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

