Breast cancer cell migration is regulated through junctional adhesion molecule-A-mediated activation of Rap1 GTPase
- PMID: 21429211
- PMCID: PMC3219194
- DOI: 10.1186/bcr2853
Breast cancer cell migration is regulated through junctional adhesion molecule-A-mediated activation of Rap1 GTPase
Abstract
Introduction: The adhesion protein junctional adhesion molecule-A (JAM-A) regulates epithelial cell morphology and migration, and its over-expression has recently been linked with increased risk of metastasis in breast cancer patients. As cell migration is an early requirement for tumor metastasis, we sought to identify the JAM-A signalling events regulating migration in breast cancer cells.
Methods: MCF7 breast cancer cells (which express high endogenous levels of JAM-A) and primary cultures from breast cancer patients were used for this study. JAM-A was knocked down in MCF7 cells using siRNA to determine the consequences for cell adhesion, cell migration and the protein expression of various integrin subunits. As we had previously demonstrated a link between the expression of JAM-A and β1-integrin, we examined activation of the β1-integrin regulator Rap1 GTPase in response to JAM-A knockdown or functional antagonism. To test whether JAM-A, Rap1 and β1-integrin lie in a linear pathway, we tested functional inhibitors of all three proteins separately or together in migration assays. Finally we performed immunoprecipitations in MCF7 cells and primary breast cells to determine the binding partners connecting JAM-A to Rap1 activation.
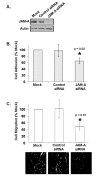
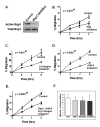
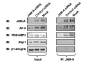
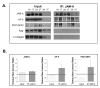
Results: JAM-A knockdown in MCF7 breast cancer cells reduced adhesion to, and migration through, the β1-integrin substrate fibronectin. This was accompanied by reduced protein expression of β1-integrin and its binding partners αV- and α5-integrin. Rap1 activity was reduced in response to JAM-A knockdown or inhibition, and pharmacological inhibition of Rap1 reduced MCF7 cell migration. No additive anti-migratory effect was observed in response to simultaneous inhibition of JAM-A, Rap1 and β1-integrin, suggesting that they lie in a linear migratory pathway. Finally, in an attempt to elucidate the binding partners putatively linking JAM-A to Rap1 activation, we have demonstrated the formation of a complex between JAM-A, AF-6 and the Rap1 activator PDZ-GEF2 in MCF7 cells and in primary cultures from breast cancer patients.
Conclusions: Our findings provide compelling evidence of a novel role for JAM-A in driving breast cancer cell migration via activation of Rap1 GTPase and β1-integrin. We speculate that JAM-A over-expression in some breast cancer patients may represent a novel therapeutic target to reduce the likelihood of metastasis.
Figures


References
-
- Hough CD, Sherman-Baust CA, Pizer ES, Montz FJ, Im DD, Rosenshein NB, Cho KR, Riggins GJ, Morin PJ. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res. 2000;60:6281–6287. - PubMed
-
- Kominsky SL, Argani P, Korz D, Evron E, Raman V, Garrett E, Rein A, Sauter G, Kallioniemi OP, Sukumar S. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene. 2003;22:2021–2033. doi: 10.1038/sj.onc.1206199. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

