Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis
- PMID: 21430186
- PMCID: PMC3091056
- DOI: 10.1104/pp.111.172684
Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis
Abstract
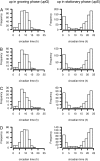
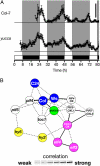
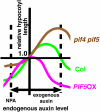
Plants exhibit daily rhythms in their growth, providing an ideal system for the study of interactions between environmental stimuli such as light and internal regulators such as the circadian clock. We previously found that two basic loop-helix-loop transcription factors, PHYTOCHROME-INTERACTING FACTOR4 (PIF4) and PIF5, integrate light and circadian clock signaling to generate rhythmic plant growth in Arabidopsis (Arabidopsis thaliana). Here, we use expression profiling and real-time growth assays to identify growth regulatory networks downstream of PIF4 and PIF5. Genome-wide analysis of light-, clock-, or growth-correlated genes showed significant overlap between the transcriptomes of clock-, light-, and growth-related pathways. Overrepresentation analysis of growth-correlated genes predicted that the auxin and gibberellic acid (GA) hormone pathways both contribute to diurnal growth control. Indeed, lesions of GA biosynthesis genes retarded rhythmic growth. Surprisingly, GA-responsive genes are not enriched among genes regulated by PIF4 and PIF5, whereas auxin pathway and response genes are. Consistent with this finding, the auxin response is more severely affected than the GA response in pif4 pif5 double mutants and in PIF5-overexpressing lines. We conclude that at least two downstream modules participate in diurnal rhythmic hypocotyl growth: PIF4 and/or PIF5 modulation of auxin-related pathways and PIF-independent regulation of the GA pathway.
Figures




References
-
- Alabadí D, Blázquez MA. (2009) Molecular interactions between light and hormone signaling to control plant growth. Plant Mol Biol 69: 409–417 - PubMed
-
- Anastasiou E, Lenhard M. (2007) Growing up to one’s standard. Curr Opin Plant Biol 10: 63–69 - PubMed
-
- Bae G, Choi G. (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59: 281–311 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

