MarR homologs with urate-binding signature
- PMID: 21432936
- PMCID: PMC3064840
- DOI: 10.1002/pro.588
MarR homologs with urate-binding signature
Abstract
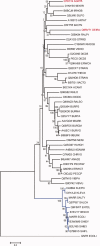
Bacteria that associate with living hosts require intricate mechanisms to detect and respond to host defenses. Part of the early host defense against invading bacteria is the production of reactive oxygen species, and xanthine oxidase is one of the main producers of such agents. The end-product of the enzymatic activity of xanthine oxidase, urate, was previously shown to be the natural ligand for Deinococcus radiodurans-encoded HucR and it was shown to attenuate DNA binding by Agrobacterium tumefaciens-encoded PecS and Burkholderia thailandensis-encoded MftR, all members of the multiple antibiotic resistance regulator (MarR) family. We here show that residues involved in binding urate and eliciting the DNA binding antagonism are conserved in a specific subset of MarR homologs. Although HucR controls endogenous urate levels by regulating a uricase gene, almost all other homologs are predicted to respond to exogenous urate levels and to regulate a transmembrane transport protein belonging to either the drug metabolite transporter (DMT) or the major facilitator superfamily (MFS), as further evidenced by the presence of conserved binding sites for the cognate transcription factor within the respective promoter regions. These data suggest the use of orthologous genes for different regulatory purposes. We propose the designation UrtR (urate responsive transcriptional regulator) for this distinct subfamily of MarR homologs based on their common mechanism of urate-mediated attenuation of DNA binding.
Copyright © 2011 The Protein Society.
Figures
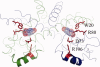


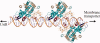
Similar articles
-
Ligand-binding pocket bridges DNA-binding and dimerization domains of the urate-responsive MarR homologue MftR from Burkholderia thailandensis.Biochemistry. 2014 Jul 15;53(27):4368-80. doi: 10.1021/bi500219t. Epub 2014 Jul 1. Biochemistry. 2014. PMID: 24955985 Free PMC article.
-
Urate-responsive MarR homologs from Burkholderia.Mol Biosyst. 2010 Nov;6(11):2133-42. doi: 10.1039/c0mb00086h. Epub 2010 Aug 23. Mol Biosyst. 2010. PMID: 20730247
-
HucR, a novel uric acid-responsive member of the MarR family of transcriptional regulators from Deinococcus radiodurans.J Biol Chem. 2004 Dec 3;279(49):51442-50. doi: 10.1074/jbc.M405586200. Epub 2004 Sep 24. J Biol Chem. 2004. PMID: 15448166
-
Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins.Curr Issues Mol Biol. 2006 Jan;8(1):51-62. Curr Issues Mol Biol. 2006. PMID: 16450885 Review.
-
MarR family transcription factors: dynamic variations on a common scaffold.Crit Rev Biochem Mol Biol. 2017 Dec;52(6):595-613. doi: 10.1080/10409238.2017.1344612. Epub 2017 Jul 3. Crit Rev Biochem Mol Biol. 2017. PMID: 28670937 Review.
Cited by
-
Transcriptional control of lipid metabolism by the MarR-like regulator FamR and the global regulator GlxR in the lipophilic axilla isolate Corynebacterium jeikeium K411.Microb Biotechnol. 2013 Mar;6(2):118-30. doi: 10.1111/1751-7915.12004. Epub 2012 Nov 20. Microb Biotechnol. 2013. PMID: 23163914 Free PMC article.
-
MarR Family Transcriptional Regulators and Their Roles in Plant-Interacting Bacteria.Microorganisms. 2023 Jul 29;11(8):1936. doi: 10.3390/microorganisms11081936. Microorganisms. 2023. PMID: 37630496 Free PMC article. Review.
-
Targeting bacterial transcription factors for infection control: opportunities and challenges.Transcription. 2025 Feb;16(1):141-168. doi: 10.1080/21541264.2023.2293523. Epub 2023 Dec 21. Transcription. 2025. PMID: 38126125 Free PMC article. Review.
-
Ligand-binding pocket bridges DNA-binding and dimerization domains of the urate-responsive MarR homologue MftR from Burkholderia thailandensis.Biochemistry. 2014 Jul 15;53(27):4368-80. doi: 10.1021/bi500219t. Epub 2014 Jul 1. Biochemistry. 2014. PMID: 24955985 Free PMC article.
-
Transcription Factor PecS Mediates Agrobacterium fabrum Fitness and Survival.J Bacteriol. 2023 Jul 25;205(7):e0047822. doi: 10.1128/jb.00478-22. Epub 2023 Jun 14. J Bacteriol. 2023. PMID: 37314346 Free PMC article.
References
-
- Victor VM, Rocha M, De la Fuente M. Immune cells: free radicals and antioxidants in sepsis. Int Immunopharmacol. 2004;4:327–347. - PubMed
-
- Nanda AK, Andrio E, Marino D, Pauly N, Dunand C. Reactive oxygen species during plant-microorganism early interactions. J Integr Plant Biol. 2010;52:195–204. - PubMed
-
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. - PubMed
-
- Cox MM, Battista JR. Deinococcus radiodurans: the consummate survivor. Nat Rev Microbiol. 2005;3:882–892. - PubMed
-
- Wilkinson SP, Grove A. HucR, a novel uric acid-responsive member of the MarR family of transcriptional regulators from Deinococcus radiodurans. J Biol Chem. 2004;279:51442–51450. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

