tmRNA regulates synthesis of the ArfA ribosome rescue factor
- PMID: 21435036
- PMCID: PMC3103599
- DOI: 10.1111/j.1365-2958.2011.07638.x
tmRNA regulates synthesis of the ArfA ribosome rescue factor
Abstract
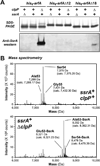
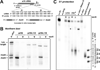
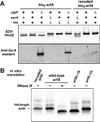
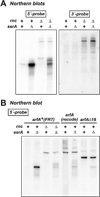
Translation of mRNA lacking an in-frame stop codon leads to ribosome arrest at the 3' end of the transcript. In bacteria, the tmRNA quality control system recycles these stalled ribosomes and tags the incomplete nascent chains for degradation. Although ubiquitous in eubacteria, the ssrA gene encoding tmRNA is not essential for the viability of Escherichia coli and other model bacterial species. ArfA (YhdL) is a mediator of tmRNA-independent ribosome rescue that is essential for the viability of E. coliΔssrA mutants. Here, we demonstrate that ArfA is synthesized from truncated mRNA and therefore regulated by tmRNA tagging activity. RNase III cleaves a hairpin structure within the arfA-coding sequence to produce transcripts that lack stop codons. In the absence of tmRNA tagging, truncated ArfA chains are released from the ribosome. The truncated ArfAΔ18 protein (which lacks 18 C-terminal residues) is functional in ribosome rescue and supports ΔssrA cell viability when expressed from the arfA locus. Other proteobacterial arfA genes also encode hairpins, and transcripts from Dickeya dadantii and Salmonella typhimurium are cleaved by RNase III when expressed in E. coli. Thus, synthesis of ArfA from truncated mRNA appears to be a general mechanism to regulate alternative ribosome rescue activity.
© 2011 Blackwell Publishing Ltd.
Figures





References
-
- Chadani Y, Ono K, Kutsukake K, Abo T. Escherichia coli YaeJ protein mediates a novel ribosome-rescue pathway distinct from SsrA- and ArfA-mediated pathways. Mol Microbiol. 2011 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

