Comparative transcriptome profiling of amyloid precursor protein family members in the adult cortex
- PMID: 21435241
- PMCID: PMC3080314
- DOI: 10.1186/1471-2164-12-160
Comparative transcriptome profiling of amyloid precursor protein family members in the adult cortex
Abstract
Background: The β-amyloid precursor protein (APP) and the related β-amyloid precursor-like proteins (APLPs) undergo complex proteolytic processing giving rise to several fragments. Whereas it is well established that Aβ accumulation is a central trigger for Alzheimer's disease, the physiological role of APP family members and their diverse proteolytic products is still largely unknown. The secreted APPsα ectodomain has been shown to be involved in neuroprotection and synaptic plasticity. The γ-secretase-generated APP intracellular domain (AICD) functions as a transcriptional regulator in heterologous reporter assays although its role for endogenous gene regulation has remained controversial.
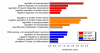
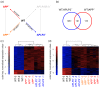
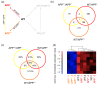
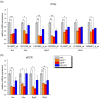
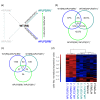
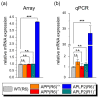
Results: To gain further insight into the molecular changes associated with knockout phenotypes and to elucidate the physiological functions of APP family members including their proposed role as transcriptional regulators, we performed DNA microarray transcriptome profiling of prefrontal cortex of adult wild-type (WT), APP knockout (APP-/-), APLP2 knockout (APLP2-/-) and APPsα knockin mice (APPα/α) expressing solely the secreted APPsα ectodomain. Biological pathways affected by the lack of APP family members included neurogenesis, transcription, and kinase activity. Comparative analysis of transcriptome changes between mutant and wild-type mice, followed by qPCR validation, identified co-regulated gene sets. Interestingly, these included heat shock proteins and plasticity-related genes that were both down-regulated in knockout cortices. In contrast, we failed to detect significant differences in expression of previously proposed AICD target genes including Bace1, Kai1, Gsk3b, p53, Tip60, and Vglut2. Only Egfr was slightly up-regulated in APLP2-/- mice. Comparison of APP-/- and APPα/α with wild-type mice revealed a high proportion of co-regulated genes indicating an important role of the C-terminus for cellular signaling. Finally, comparison of APLP2-/- on different genetic backgrounds revealed that background-related transcriptome changes may dominate over changes due to the knockout of a single gene.
Conclusion: Shared transcriptome profiles corroborated closely related physiological functions of APP family members in the adult central nervous system. As expression of proposed AICD target genes was not altered in adult cortex, this may indicate that these genes are not affected by lack of APP under resting conditions or only in a small subset of cells.
Figures
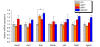
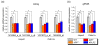

References
-
- Kimberly WT, Zheng JB, Guenette SY, Selkoe DJ. The intracellular domain of the beta-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J Biol Chem. 2001;276:40288–40292. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

