Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signaling
- PMID: 21435554
- PMCID: PMC3383999
- DOI: 10.1016/j.neuron.2011.02.018
Proneural transcription factors regulate different steps of cortical neuron migration through Rnd-mediated inhibition of RhoA signaling
Abstract
Little is known of the intracellular machinery that controls the motility of newborn neurons. We have previously shown that the proneural protein Neurog2 promotes the migration of nascent cortical neurons by inducing the expression of the atypical Rho GTPase Rnd2. Here, we show that another proneural factor, Ascl1, promotes neuronal migration in the cortex through direct regulation of a second Rnd family member, Rnd3. Both Rnd2 and Rnd3 promote neuronal migration by inhibiting RhoA signaling, but they control distinct steps of the migratory process, multipolar to bipolar transition in the intermediate zone and locomotion in the cortical plate, respectively. Interestingly, these divergent functions directly result from the distinct subcellular distributions of the two Rnd proteins. Because Rnd proteins also regulate progenitor divisions and neurite outgrowth, we propose that proneural factors, through spatiotemporal regulation of Rnd proteins, integrate the process of neuronal migration with other events in the neurogenic program.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
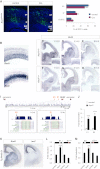
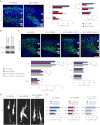

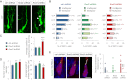
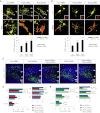


Comment in
-
Rnd-ing up RhoA activity to link neurogenesis with steps in neuronal migration.Dev Cell. 2011 Apr 19;20(4):409-10. doi: 10.1016/j.devcel.2011.04.001. Dev Cell. 2011. PMID: 21497751
References
-
- Armentano M., Filosa A., Andolfi G., Studer M. COUP-TFI is required for the formation of commissural projections in the forebrain by regulating axonal growth. Development. 2006;133:4151–4162. - PubMed
-
- Bektic J., Pfeil K., Berger A.P., Ramoner R., Pelzer A., Schäfer G., Kofler K., Bartsch G., Klocker H. Small G-protein RhoE is underexpressed in prostate cancer and induces cell cycle arrest and apoptosis. Prostate. 2005;64:332–340. - PubMed
-
- Britz O., Mattar P., Nguyen L., Langevin L.M., Zimmer C., Alam S., Guillemot F., Schuurmans C. A role for proneural genes in the maturation of cortical progenitor cells. Cereb.Cortex. 2006;16(Suppl1 1):i138–i151. - PubMed
-
- Castro D.S., Skowronska-Krawczyk D., Armant O., Donaldson I.J., Parras C., Hunt C., Critchley J.A., Nguyen L., Gossler A., Göttgens B. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev. Cell. 2006;11:831–844. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_U117570528/MRC_/Medical Research Council/United Kingdom
- 086947/Z/08/Z/WT_/Wellcome Trust/United Kingdom
- U117570528/MRC_/Medical Research Council/United Kingdom
- BB/E004083/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/E004083/2/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

