RUNX transcription factor-mediated association of Cd4 and Cd8 enables coordinate gene regulation
- PMID: 21435585
- PMCID: PMC3101577
- DOI: 10.1016/j.immuni.2011.03.004
RUNX transcription factor-mediated association of Cd4 and Cd8 enables coordinate gene regulation
Abstract
T cell fate is associated with mutually exclusive expression of CD4 or CD8 in helper and cytotoxic T cells, respectively. How expression of one locus is temporally coordinated with repression of the other has been a long-standing enigma, though we know RUNX transcription factors activate the Cd8 locus, silence the Cd4 locus, and repress the Zbtb7b locus (encoding the transcription factor ThPOK), which is required for CD4 expression. Here we found that nuclear organization was altered by interplay among members of this transcription factor circuitry: RUNX binding mediated association of Cd4 and Cd8 whereas ThPOK binding kept the loci apart. Moreover, targeted deletions within Cd4 modulated CD8 expression and pericentromeric repositioning of Cd8. Communication between Cd4 and Cd8 thus appears to enable long-range epigenetic regulation to ensure that expression of one excludes the other in mature CD4 or CD8 single-positive (SP) cells.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
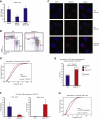
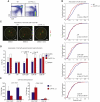


References
-
- Bacher C.P., Guggiari M., Brors B., Augui S., Clerc P., Avner P., Eils R., Heard E. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 2006;8:293–299. - PubMed
-
- Brown K.E., Baxter J., Graf D., Merkenschlager M., Fisher A.G. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell. 1999;3:207–217. - PubMed
-
- Campbell R. Cambridge University Press; Cambridge, UK: 1989. Statistics for Biologists.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

