Estimate of effective recombination rate and average selection coefficient for HIV in chronic infection
- PMID: 21436045
- PMCID: PMC3078368
- DOI: 10.1073/pnas.1102036108
Estimate of effective recombination rate and average selection coefficient for HIV in chronic infection
Abstract
HIV adaptation to a host in chronic infection is simulated by means of a Monte-Carlo algorithm that includes the evolutionary factors of mutation, positive selection with varying strength among sites, random genetic drift, linkage, and recombination. By comparing two sensitive measures of linkage disequilibrium (LD) and the number of diverse sites measured in simulation to patient data from one-time samples of pol gene obtained by single-genome sequencing from representative untreated patients, we estimate the effective recombination rate and the average selection coefficient to be on the order of 1% per genome per generation (10(-5) per base per generation) and 0.5%, respectively. The adaptation rate is twofold higher and fourfold lower than predicted in the absence of recombination and in the limit of very frequent recombination, respectively. The level of LD and the number of diverse sites observed in data also range between the values predicted in simulation for these two limiting cases. These results demonstrate the critical importance of finite population size, linkage, and recombination in HIV evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
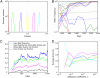
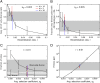
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

