Performance, accuracy, and Web server for evolutionary placement of short sequence reads under maximum likelihood
- PMID: 21436105
- PMCID: PMC3078422
- DOI: 10.1093/sysbio/syr010
Performance, accuracy, and Web server for evolutionary placement of short sequence reads under maximum likelihood
Abstract
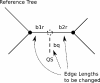
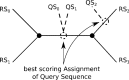
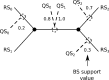
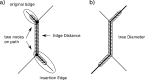
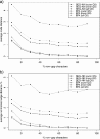
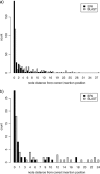
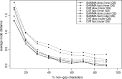
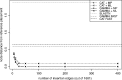
We present an evolutionary placement algorithm (EPA) and a Web server for the rapid assignment of sequence fragments (short reads) to edges of a given phylogenetic tree under the maximum-likelihood model. The accuracy of the algorithm is evaluated on several real-world data sets and compared with placement by pair-wise sequence comparison, using edit distances and BLAST. We introduce a slow and accurate as well as a fast and less accurate placement algorithm. For the slow algorithm, we develop additional heuristic techniques that yield almost the same run times as the fast version with only a small loss of accuracy. When those additional heuristics are employed, the run time of the more accurate algorithm is comparable with that of a simple BLAST search for data sets with a high number of short query sequences. Moreover, the accuracy of the EPA is significantly higher, in particular when the sample of taxa in the reference topology is sparse or inadequate. Our algorithm, which has been integrated into RAxML, therefore provides an equally fast but more accurate alternative to BLAST for tree-based inference of the evolutionary origin and composition of short sequence reads. We are also actively developing a Web server that offers a freely available service for computing read placements on trees using the EPA.
Figures

References
-
- Ababneh F, Jermiin LS, Ma C, Robinson J. Matched-pairs tests of homogeneity with applications to homologous nucleotide sequences. Bioinformatics. 2006;22:1225–1231. - PubMed
-
- Berger SA, Stamatakis A. Accuracy of morphology-based phylogenetic fossil placement under maximum likelihood. Proceedings of IEEE/ACS International Conference on Computer Systems and Applications (AICCSA-10); 2010 May 16–18; Hammamet, Tunisia: IEEE Computer Society. p. 1–8. 2010
-
- Bininda-Emonds ORP, Brady SG, Sanderson MJ, Kim J. Scaling of accuracy in extremely large phylogenetic trees. In: Altman RB, Dunker AK, Hunter L, Lauderdale K, Klein TE, editors. Pacific Symposium on Biocomputing 2001. River Edge (NJ): World Scientific; 2000. pp. 547–558. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

