An ENU-induced mutation in the Mertk gene (Mertknmf12) leads to a slow form of retinal degeneration
- PMID: 21436282
- PMCID: PMC3175976
- DOI: 10.1167/iovs.10-7077
An ENU-induced mutation in the Mertk gene (Mertknmf12) leads to a slow form of retinal degeneration
Abstract
Purpose: To determine the basis and to characterize the phenotype of a chemically induced mutation in a mouse model of retinal degeneration.
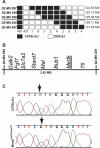
Methods: Screening by indirect ophthalmoscopy identified a line of N-ethyl-N-nitrosourea (ENU) mutagenized mice demonstrating retinal patches. Longitudinal studies of retinal histologic sections showed photoreceptors in the peripheral retina undergoing slow, progressive degeneration. The mutation was named neuroscience mutagenesis facility 12 (nmf12), and mapping localized the critical region to Chromosome 2.
Results: Sequencing of nmf12 DNA revealed a point mutation in the c-mer tyrosine kinase gene, designated Mertk(nmf12). We detected elevated levels of tumor necrosis factor (Tnf, previously Tnfa) in retinas of Mertk(nmf12) homozygotes relative to wild-type controls and investigated whether the increase of TNF, an inflammatory cytokine produced by macrophages/monocytes that signals intracellularly to cause necrosis or apoptosis, could underlie the retinal degeneration observed in Mertk(nmf12) homozygotes. Mertk(nmf12) homozygous mice were mated to mice lacking the entire Tnf gene and partial coding sequences of the Lta (Tnfb) and Ltb (Tnfc) genes.(2) B6.129P2-Ltb/Tnf/Lta(tm1Dvk)/J homozygotes did not exhibit a retinal degeneration phenotype and will, hereafter, be referred to as Tnfabc(-/-) mice. Surprisingly, mice homozygous for both the Mertk(nmf12) and the Ltb/Tnf/Lta(tm1Dvk) allele (Tnfabc(-/-)) demonstrated an increase in the rate of retinal degeneration.
Conclusions: These findings illustrate that a mutation in the Mertk gene leads to a significantly slower progressive retinal degeneration compared with other alleles of Mertk. These results demonstrate that TNF family members play a role in protecting photoreceptors of Mertk(nmf12) homozygotes from cell death.
Figures






References
-
- Marmor MF. Mechanisms of fluid accumulation in retinal edema. Doc Ophthalmol. 1993;97:179–203 - PubMed
-
- Allikmets R, Seddon JM, Bernstein PS, et al. Evaluation of the Best disease gene in patients with age-related macular degeneration and other maculopathies. Hum Genet. 1999;104:449–453 - PubMed
-
- Bakall B, Marknell T, Ingvast S, et al. The mutation spectrum of the bestrophin protein- functional implications. Hum Genet. 1999;104:338–389 - PubMed
-
- Caldwell GM, Kakuk LE, Griesinger IB, et al. Bestrophin gene mutations in patients with Best vitelliform macular dystrophy. Genomics. 1999;58:98–101 - PubMed
-
- Ambati J, Ambati BK, Yoo SH, Ianchulev S, Adamis AP. Age-related macular degeneration: etiology, pathogenesis and and therapeutic strategies. Surv Ophthalmol. 2003;48:257–293 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

