Single virus genomics: a new tool for virus discovery
- PMID: 21436882
- PMCID: PMC3059205
- DOI: 10.1371/journal.pone.0017722
Single virus genomics: a new tool for virus discovery
Abstract
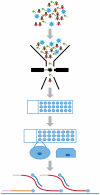
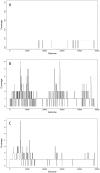
Whole genome amplification and sequencing of single microbial cells has significantly influenced genomics and microbial ecology by facilitating direct recovery of reference genome data. However, viral genomics continues to suffer due to difficulties related to the isolation and characterization of uncultivated viruses. We report here on a new approach called 'Single Virus Genomics', which enabled the isolation and complete genome sequencing of the first single virus particle. A mixed assemblage comprised of two known viruses; E. coli bacteriophages lambda and T4, were sorted using flow cytometric methods and subsequently immobilized in an agarose matrix. Genome amplification was then achieved in situ via multiple displacement amplification (MDA). The complete lambda phage genome was recovered with an average depth of coverage of approximately 437X. The isolation and genome sequencing of uncultivated viruses using Single Virus Genomics approaches will enable researchers to address questions about viral diversity, evolution, adaptation and ecology that were previously unattainable.
Conflict of interest statement
Figures


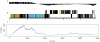
References
-
- Lasken RS. Single-cell genomic sequencing using Multiple Displacement Amplification. Curr Opin Microbiol. 2007;10(5):510–516. - PubMed
-
- Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth's biogeochemical cycles. Science. 2008;320(5879):1034–1039. - PubMed
-
- Amon RMW, Benner R. Bacterial utilization of different size classes of dissolved organic matter. Limnology and Oceanography. 1996;41(1):41–51.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

